Gene
KWMTBOMO10887
Annotation
antennal_binding_protein_4_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 1.339 Nuclear Reliability : 1.042
Sequence
CDS
ATGAGAATTAGTTTTTTGTTTTTGATTAGTGTAACTATTATTACATTTGACAGCGTTTTTGCGATGACTCGTGCCCAGGTTAAAAAAACAATGACAATCATGAAAAATCAATGTATGCCGAAAAATGGGGTTACCGAAGATCAAGTTGGGAAAATTGAAGAAGGTATCTTTCTAGAAAATCACAATGTGATGTGTTACATCGCTTGCGTTTATAAAACTATTCAAGTTGTTAAGAATGACAGACTCGATAAAGACCTGATATCGAAACAAATTGATGTATTATATCCTCAAGAAATTAGGGAATCTACCAAAAAGGCTGTTGGAGATTGCATTAATTTGCAGGAAAAATACGACGATTGGTGTGAAGGTATATTTCGCTCGACCAAATGTTTGTATGAAAAAGATCCAGCAAACTTTATTTTTCCTTAG
Protein
MRISFLFLISVTIITFDSVFAMTRAQVKKTMTIMKNQCMPKNGVTEDQVGKIEEGIFLENHNVMCYIACVYKTIQVVKNDRLDKDLISKQIDVLYPQEIRESTKKAVGDCINLQEKYDDWCEGIFRSTKCLYEKDPANFIFP
Summary
Uniprot
A0A212EMX0
A0A1Z2R8P1
A0A2K8GKK4
A0A076E5R4
S4WVB2
A0A2H1W236
+ More
A0A1V1WC74 A0A0K8TW06 A0A0X9ND42 A0A0G2YHW6 A0A173ACX8 H9XFD2 A0A1B3P5H2 A0A1B3P5I2 A0A2R4SBB1 A0A3G2YUX0 A0A0K8TUS9 X2CDC6 A0A0X9NBU7 Q8WRW8 A0A194PH26 A0A2H4FXX5 A0A194QLX3 A0A3S2TIL0 A0A0M4JHK6 A0A345BES8 A0A1B4ZBK3 H9N4R9 A0A146JYP8 A0A2H1W3L4 A0A146JYK6 A0A1V1WC69 H9JFQ4 A0A3G6V7H4 A0A1B3P5I8 S5TP47 A0A0K8TVF6 A0A386H951 A0A0M4FJN0 A0A1L2BLC6 Q8WRW7 A0A0K8TUU0 A0A0M5J840 A0A345BES7 M4Y1X0 A0A3G2YV88 H9JFQ5 A0A0G2YHX1 B8ZWK1 A0A2H1W1X3 M4TBS8 A0A109YJX5 A0A1B4ZBM4 H9N4S1 A0A1V1WCH5 A0A1V1WBZ5 A0A2K8GKL4 F5ANI0 A0A2W1BM18 D2SNM0 A0A194QMH1 A0A1B4ZBK2 A0A194PIR2 A0A194PHT0 A0A109XL66 A0A386H8W3 G9I569 A0A0P0E082 A0A212EMZ7 A0A2A4K884 X2C512 A0A076E7D5 A0A076E937 S5T764 A0A1L2BLE2 A0A2H1W2M5 S4X0Q1 A0A2R4SBE0 A0A0M5KLD9 A0A2W1BRD6 A0A212EN23 A0A076E5N5 A0A1L2BLA9 A0A3G2YUY2 A0A2A4K7Q1 A0A386H970 H9N4R6 A0A0K8TUB9 A0A1B3P5J9 A0A3G2YV58 A0A194PGT4 A0A2R4SBD2 A0A2W1BKB0 A0A2A4K934 F5ANI3 A0A345BER0 A0A2K8GL24 A0A3G6V623
A0A1V1WC74 A0A0K8TW06 A0A0X9ND42 A0A0G2YHW6 A0A173ACX8 H9XFD2 A0A1B3P5H2 A0A1B3P5I2 A0A2R4SBB1 A0A3G2YUX0 A0A0K8TUS9 X2CDC6 A0A0X9NBU7 Q8WRW8 A0A194PH26 A0A2H4FXX5 A0A194QLX3 A0A3S2TIL0 A0A0M4JHK6 A0A345BES8 A0A1B4ZBK3 H9N4R9 A0A146JYP8 A0A2H1W3L4 A0A146JYK6 A0A1V1WC69 H9JFQ4 A0A3G6V7H4 A0A1B3P5I8 S5TP47 A0A0K8TVF6 A0A386H951 A0A0M4FJN0 A0A1L2BLC6 Q8WRW7 A0A0K8TUU0 A0A0M5J840 A0A345BES7 M4Y1X0 A0A3G2YV88 H9JFQ5 A0A0G2YHX1 B8ZWK1 A0A2H1W1X3 M4TBS8 A0A109YJX5 A0A1B4ZBM4 H9N4S1 A0A1V1WCH5 A0A1V1WBZ5 A0A2K8GKL4 F5ANI0 A0A2W1BM18 D2SNM0 A0A194QMH1 A0A1B4ZBK2 A0A194PIR2 A0A194PHT0 A0A109XL66 A0A386H8W3 G9I569 A0A0P0E082 A0A212EMZ7 A0A2A4K884 X2C512 A0A076E7D5 A0A076E937 S5T764 A0A1L2BLE2 A0A2H1W2M5 S4X0Q1 A0A2R4SBE0 A0A0M5KLD9 A0A2W1BRD6 A0A212EN23 A0A076E5N5 A0A1L2BLA9 A0A3G2YUY2 A0A2A4K7Q1 A0A386H970 H9N4R6 A0A0K8TUB9 A0A1B3P5J9 A0A3G2YV58 A0A194PGT4 A0A2R4SBD2 A0A2W1BKB0 A0A2A4K934 F5ANI3 A0A345BER0 A0A2K8GL24 A0A3G6V623
Pubmed
EMBL
AGBW02013764
OWR42850.1
KY810183
ASA40075.1
KX585270
ARO70163.1
+ More
KF487603 AII01001.1 KC817142 AGP03456.1 ODYU01005815 SOQ47083.1 GENK01000017 JAV45896.1 GCVX01000005 JAI18225.1 KT220705 ALZ45423.1 KP331517 AKI87963.1 KT156677 ANG08530.1 JN867059 AFG72998.1 KX655909 AOG12858.1 KX655919 AOG12868.1 MF066357 AVZ44704.1 MG820723 AYP30815.1 GCVX01000006 JAI18224.1 KC763848 AGM38605.1 KT220702 ALZ45420.1 AF393490 AAL60415.1 KQ459604 KPI92602.1 KY130466 APG32534.1 KQ461196 KPJ06553.1 RSAL01000113 RVE47000.1 KT192038 ALD65891.1 MG788234 AXF48752.1 LC027692 BAV56795.1 JF720860 AFD34178.1 GEDO01000010 JAP88616.1 SOQ47084.1 GEDO01000022 JAP88604.1 GENK01000027 JAV45886.1 BABH01001858 MH027974 AZB49392.1 KX655921 AOG12870.1 KC887521 AGS36755.1 GCVX01000004 JAI18226.1 MG546562 AYD42185.1 KP985225 ALC76547.1 KT327218 ALS03861.1 AF393491 AAL60416.1 GCVX01000003 JAI18227.1 KP985223 GEDO01000015 ALC76545.1 JAP88611.1 MG788233 AXF48751.1 KC507183 AGI37366.1 MG820726 AYP30818.1 BABH01001857 KP331522 KT261669 AKI87968.1 ALJ30210.1 FM876234 CAS90125.1 SOQ47081.1 JX962790 AGH70103.1 KT220704 ALZ45422.1 LC027695 BAV56798.1 JF720862 AFD34180.1 GENK01000022 JAV45891.1 GENK01000021 JAV45892.1 KX585267 ARO70160.1 HQ436363 AEB54586.1 KZ149999 PZC75331.1 EZ407149 ACX53711.1 KPJ06554.1 LC027686 BAV56789.1 KPI92604.1 KPI92603.1 KT220706 ALZ45424.1 MG546555 AYD42178.1 JN571540 AEX07275.1 KT261656 ALJ30197.1 OWR42849.1 NWSH01000044 PCG80279.1 KC763856 AGM38613.1 KF487599 AII00997.1 KF487591 AII00989.1 KC887511 AGS36747.1 KT327216 ALS03859.1 SOQ47082.1 KC817144 AGP03458.1 MG581971 AVZ44714.1 KT192036 ALD65889.1 PZC75330.1 OWR42851.1 KF487573 AII00971.1 KT327220 ALS03863.1 MG820718 AYP30810.1 PCG80305.1 MG546570 AYD42193.1 JF720857 AFD34175.1 GCVX01000153 JAI18077.1 KX655924 AOG12873.1 MG820730 AYP30822.1 KPI92601.1 MF066360 AVZ44707.1 PZC75328.1 PCG80303.1 HQ436366 AEB54589.1 MG788216 AXF48734.1 KY225481 ARO70493.1 MH027965 AZB49383.1
KF487603 AII01001.1 KC817142 AGP03456.1 ODYU01005815 SOQ47083.1 GENK01000017 JAV45896.1 GCVX01000005 JAI18225.1 KT220705 ALZ45423.1 KP331517 AKI87963.1 KT156677 ANG08530.1 JN867059 AFG72998.1 KX655909 AOG12858.1 KX655919 AOG12868.1 MF066357 AVZ44704.1 MG820723 AYP30815.1 GCVX01000006 JAI18224.1 KC763848 AGM38605.1 KT220702 ALZ45420.1 AF393490 AAL60415.1 KQ459604 KPI92602.1 KY130466 APG32534.1 KQ461196 KPJ06553.1 RSAL01000113 RVE47000.1 KT192038 ALD65891.1 MG788234 AXF48752.1 LC027692 BAV56795.1 JF720860 AFD34178.1 GEDO01000010 JAP88616.1 SOQ47084.1 GEDO01000022 JAP88604.1 GENK01000027 JAV45886.1 BABH01001858 MH027974 AZB49392.1 KX655921 AOG12870.1 KC887521 AGS36755.1 GCVX01000004 JAI18226.1 MG546562 AYD42185.1 KP985225 ALC76547.1 KT327218 ALS03861.1 AF393491 AAL60416.1 GCVX01000003 JAI18227.1 KP985223 GEDO01000015 ALC76545.1 JAP88611.1 MG788233 AXF48751.1 KC507183 AGI37366.1 MG820726 AYP30818.1 BABH01001857 KP331522 KT261669 AKI87968.1 ALJ30210.1 FM876234 CAS90125.1 SOQ47081.1 JX962790 AGH70103.1 KT220704 ALZ45422.1 LC027695 BAV56798.1 JF720862 AFD34180.1 GENK01000022 JAV45891.1 GENK01000021 JAV45892.1 KX585267 ARO70160.1 HQ436363 AEB54586.1 KZ149999 PZC75331.1 EZ407149 ACX53711.1 KPJ06554.1 LC027686 BAV56789.1 KPI92604.1 KPI92603.1 KT220706 ALZ45424.1 MG546555 AYD42178.1 JN571540 AEX07275.1 KT261656 ALJ30197.1 OWR42849.1 NWSH01000044 PCG80279.1 KC763856 AGM38613.1 KF487599 AII00997.1 KF487591 AII00989.1 KC887511 AGS36747.1 KT327216 ALS03859.1 SOQ47082.1 KC817144 AGP03458.1 MG581971 AVZ44714.1 KT192036 ALD65889.1 PZC75330.1 OWR42851.1 KF487573 AII00971.1 KT327220 ALS03863.1 MG820718 AYP30810.1 PCG80305.1 MG546570 AYD42193.1 JF720857 AFD34175.1 GCVX01000153 JAI18077.1 KX655924 AOG12873.1 MG820730 AYP30822.1 KPI92601.1 MF066360 AVZ44707.1 PZC75328.1 PCG80303.1 HQ436366 AEB54589.1 MG788216 AXF48734.1 KY225481 ARO70493.1 MH027965 AZB49383.1
Proteomes
Pfam
PF01395 PBP_GOBP
SUPFAM
SSF47565
SSF47565
Gene 3D
ProteinModelPortal
A0A212EMX0
A0A1Z2R8P1
A0A2K8GKK4
A0A076E5R4
S4WVB2
A0A2H1W236
+ More
A0A1V1WC74 A0A0K8TW06 A0A0X9ND42 A0A0G2YHW6 A0A173ACX8 H9XFD2 A0A1B3P5H2 A0A1B3P5I2 A0A2R4SBB1 A0A3G2YUX0 A0A0K8TUS9 X2CDC6 A0A0X9NBU7 Q8WRW8 A0A194PH26 A0A2H4FXX5 A0A194QLX3 A0A3S2TIL0 A0A0M4JHK6 A0A345BES8 A0A1B4ZBK3 H9N4R9 A0A146JYP8 A0A2H1W3L4 A0A146JYK6 A0A1V1WC69 H9JFQ4 A0A3G6V7H4 A0A1B3P5I8 S5TP47 A0A0K8TVF6 A0A386H951 A0A0M4FJN0 A0A1L2BLC6 Q8WRW7 A0A0K8TUU0 A0A0M5J840 A0A345BES7 M4Y1X0 A0A3G2YV88 H9JFQ5 A0A0G2YHX1 B8ZWK1 A0A2H1W1X3 M4TBS8 A0A109YJX5 A0A1B4ZBM4 H9N4S1 A0A1V1WCH5 A0A1V1WBZ5 A0A2K8GKL4 F5ANI0 A0A2W1BM18 D2SNM0 A0A194QMH1 A0A1B4ZBK2 A0A194PIR2 A0A194PHT0 A0A109XL66 A0A386H8W3 G9I569 A0A0P0E082 A0A212EMZ7 A0A2A4K884 X2C512 A0A076E7D5 A0A076E937 S5T764 A0A1L2BLE2 A0A2H1W2M5 S4X0Q1 A0A2R4SBE0 A0A0M5KLD9 A0A2W1BRD6 A0A212EN23 A0A076E5N5 A0A1L2BLA9 A0A3G2YUY2 A0A2A4K7Q1 A0A386H970 H9N4R6 A0A0K8TUB9 A0A1B3P5J9 A0A3G2YV58 A0A194PGT4 A0A2R4SBD2 A0A2W1BKB0 A0A2A4K934 F5ANI3 A0A345BER0 A0A2K8GL24 A0A3G6V623
A0A1V1WC74 A0A0K8TW06 A0A0X9ND42 A0A0G2YHW6 A0A173ACX8 H9XFD2 A0A1B3P5H2 A0A1B3P5I2 A0A2R4SBB1 A0A3G2YUX0 A0A0K8TUS9 X2CDC6 A0A0X9NBU7 Q8WRW8 A0A194PH26 A0A2H4FXX5 A0A194QLX3 A0A3S2TIL0 A0A0M4JHK6 A0A345BES8 A0A1B4ZBK3 H9N4R9 A0A146JYP8 A0A2H1W3L4 A0A146JYK6 A0A1V1WC69 H9JFQ4 A0A3G6V7H4 A0A1B3P5I8 S5TP47 A0A0K8TVF6 A0A386H951 A0A0M4FJN0 A0A1L2BLC6 Q8WRW7 A0A0K8TUU0 A0A0M5J840 A0A345BES7 M4Y1X0 A0A3G2YV88 H9JFQ5 A0A0G2YHX1 B8ZWK1 A0A2H1W1X3 M4TBS8 A0A109YJX5 A0A1B4ZBM4 H9N4S1 A0A1V1WCH5 A0A1V1WBZ5 A0A2K8GKL4 F5ANI0 A0A2W1BM18 D2SNM0 A0A194QMH1 A0A1B4ZBK2 A0A194PIR2 A0A194PHT0 A0A109XL66 A0A386H8W3 G9I569 A0A0P0E082 A0A212EMZ7 A0A2A4K884 X2C512 A0A076E7D5 A0A076E937 S5T764 A0A1L2BLE2 A0A2H1W2M5 S4X0Q1 A0A2R4SBE0 A0A0M5KLD9 A0A2W1BRD6 A0A212EN23 A0A076E5N5 A0A1L2BLA9 A0A3G2YUY2 A0A2A4K7Q1 A0A386H970 H9N4R6 A0A0K8TUB9 A0A1B3P5J9 A0A3G2YV58 A0A194PGT4 A0A2R4SBD2 A0A2W1BKB0 A0A2A4K934 F5ANI3 A0A345BER0 A0A2K8GL24 A0A3G6V623
PDB
3R72
E-value=2.55843e-14,
Score=185
Ontologies
GO
Topology
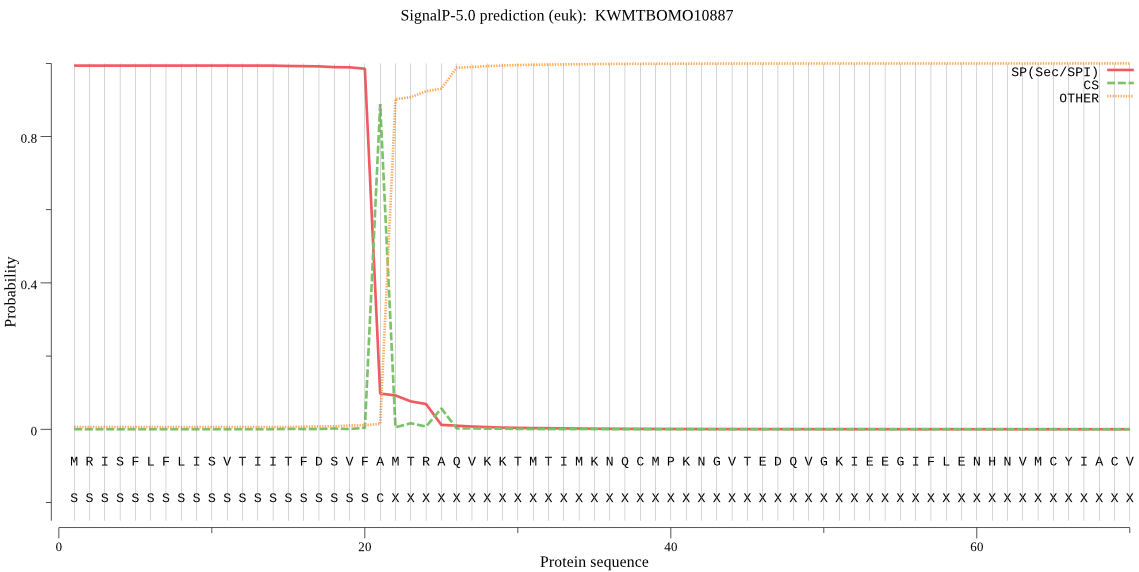
SignalP
Position: 1 - 21,
Likelihood: 0.993590
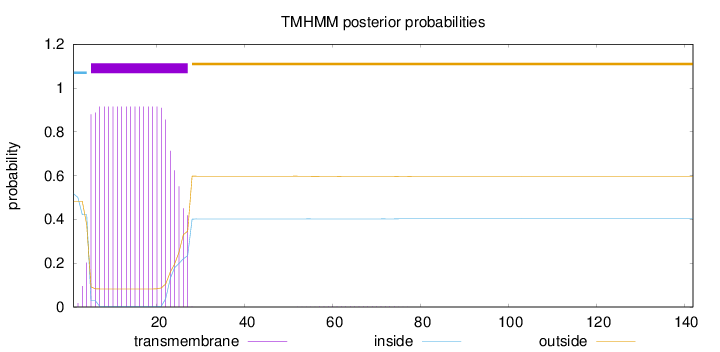
Length:
142
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.46081
Exp number, first 60 AAs:
19.43634
Total prob of N-in:
0.51739
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 142
Population Genetic Test Statistics
Pi
285.356048
Theta
203.805936
Tajima's D
1.002706
CLR
0
CSRT
0.657667116644168
Interpretation
Uncertain
