Gene
KWMTBOMO10872
Pre Gene Modal
BGIBMGA008471
Annotation
PREDICTED:_venom_protease_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.998
Sequence
CDS
ATGGGTGTAATGATACGTGTTCTGTTTTTGATGTGCATTTTGGAATGGTCCGTTTTAGCGCAATTCGGCAGTATATCAGTGAGTGTTGGCATGGGAGGAATTAGAAGCGGCAATGAAAATCTAAGCGTGAGAGCACCTTGTCCACCGAATACATCTTGTGTGCCAATCAGCTCGTGTCCGATGCTTGAAGATCTACTCGATTTCTCGTGCTTTTCATCGGATAAATACTTTCATCGTTTGAATCAACTAACGTGTGGTAATGACAACAACGAAGACTACGTTTGCTGTCCGTCGTGTGACTGCGGCCGAGTCTACCAGGAAGGCACGCAATCGTGTGGCGAGAGTATGGTACGAGGAGTTGATTATGACGGAATCGGCACACATCCTTGGGTGGCCAGAATTGGCTTCACTAAAGAAGAAACGGGTAATGTTAGATTCGCTTGCAGTGGATCTATAATATCGAAGCGAGTTGTATTAACTGCGGCGCACTGTGCTTTGGCCAAACCAGAAAGATACAAATTGTCTACGGTGGTAGTGGGCGAATGGGATATTGGACGAAGCCCCGACTGTAACGATTTCTTCTGCGCCCCTCCCACGCAAGCAATAAAAGTTGAAAGCGTTGTTGTGCACCCTGGTTACGAGGAAAAGATTTTCCGGCACGACATTGCACTTATCTTGTTGAAAGATGAAATCAAGTATTCAGTGACAGCAGCACCAGTTTGCCTGAATGAAAAACCGGAAGTTGTAATAAACGAACGCGCATCACTGGTCGGTTGGGGGAAACTGTCCGGTCAGACTAATGTGGTTAGCCGCCAGCAGCTACTAGAAGTGCCTCTTGTTCCTCTAGAAAAATGTGAAAAAGTATTTGGCGAATCCGTGCCAGTACACGAGGGGCAGCTGTGTGCGGGCGGTGAAGAAGGAAAGGATGCCTGTTCTGGATTCGGAGGAGCACCACTACTCATCAAACGTGACGGAAAATTTCTTCAGACGGGAATCGTGTCATTTGGCTCCGAAAACTGCGGCAGCGACGGCGTACCCAGCGTGTATACAAACATTGCTCATTATTATCGATGGATCGTAGAAAACTCACCCTCTTAA
Protein
MGVMIRVLFLMCILEWSVLAQFGSISVSVGMGGIRSGNENLSVRAPCPPNTSCVPISSCPMLEDLLDFSCFSSDKYFHRLNQLTCGNDNNEDYVCCPSCDCGRVYQEGTQSCGESMVRGVDYDGIGTHPWVARIGFTKEETGNVRFACSGSIISKRVVLTAAHCALAKPERYKLSTVVVGEWDIGRSPDCNDFFCAPPTQAIKVESVVVHPGYEEKIFRHDIALILLKDEIKYSVTAAPVCLNEKPEVVINERASLVGWGKLSGQTNVVSRQQLLEVPLVPLEKCEKVFGESVPVHEGQLCAGGEEGKDACSGFGGAPLLIKRDGKFLQTGIVSFGSENCGSDGVPSVYTNIAHYYRWIVENSPS
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JG24
A0A2H1V3S7
A0A212EMW9
A0A2A4J7W5
A0A194PH36
A0A2W1BMA8
+ More
A0A194QLY6 G9F9H5 N6TD06 U4UR11 A0A139WEJ1 A0A067RAE5 A0A1W4X8T0 A0A1B6DLX7 A0A1B6CAJ6 A0A2P8Z7P1 A0A1Y1LL09 A0A182WW36 A0A182UVC0 A0A1S4H1C9 A0A182L6D4 A0A182KB17 A0A1Y1LIX7 Q7PVJ0 A0A182YDQ8 A0A182U9R2 A0A1J1J398 A0A1J1ISM1 A0A182JCH5 A0A182NU45 A0A182M604 A0A1B0GLG1 W5JC77 A0A182RIP5 A0A182HGH5 A0A2M4CRS5 A0A1Y1LML9 W8B7Y4 A0A3F2YQ98 B4N0X2 Q8SXE1 B4Q9G0 B4PAG5 B4I5R0 A0A034UZP8 A0A0K8V8U7 A0A0J9R4E3 B3MNQ2 A0A0R3P0F0 A0A0A1WV38 D0QWC2 A0A1W4W8K3 Q29K28 W8B3K9 B4GXD3 A0A1I8PM58 A0A3B0JYM3 A0A1I8MF00 B3NM97 Q179F8 A0A182PE86 A0A1S4FB72 A0A2R7X3P5 A0A182VUG5 A0A1A9W0P4 A0A0Q9WBS0 B4M9B5 B4JCK5 A0A0Q9W9R8 A0A1A9XZQ4 A0A1B0BLC9 A0A084WG00 B4KHK8 A0A0M4EAX3 A0A1B0A005 A0A1A9V1F2 A0A336KWB3 A0A336N553 A0A182H2A3 A0A0Q9X4W1 A0A158NPL0 E9IDZ5 A0A195ETC0 A0A195DRH3 A0A151IIN2 A0A1B0D181 A0A026W1D1 A0A0M8ZVR1 A0A151X275 A0A154PQG6 A0A0L7QP79 A0A3L8DF44 A0A2M4BS19 A0A2M4BS12
A0A194QLY6 G9F9H5 N6TD06 U4UR11 A0A139WEJ1 A0A067RAE5 A0A1W4X8T0 A0A1B6DLX7 A0A1B6CAJ6 A0A2P8Z7P1 A0A1Y1LL09 A0A182WW36 A0A182UVC0 A0A1S4H1C9 A0A182L6D4 A0A182KB17 A0A1Y1LIX7 Q7PVJ0 A0A182YDQ8 A0A182U9R2 A0A1J1J398 A0A1J1ISM1 A0A182JCH5 A0A182NU45 A0A182M604 A0A1B0GLG1 W5JC77 A0A182RIP5 A0A182HGH5 A0A2M4CRS5 A0A1Y1LML9 W8B7Y4 A0A3F2YQ98 B4N0X2 Q8SXE1 B4Q9G0 B4PAG5 B4I5R0 A0A034UZP8 A0A0K8V8U7 A0A0J9R4E3 B3MNQ2 A0A0R3P0F0 A0A0A1WV38 D0QWC2 A0A1W4W8K3 Q29K28 W8B3K9 B4GXD3 A0A1I8PM58 A0A3B0JYM3 A0A1I8MF00 B3NM97 Q179F8 A0A182PE86 A0A1S4FB72 A0A2R7X3P5 A0A182VUG5 A0A1A9W0P4 A0A0Q9WBS0 B4M9B5 B4JCK5 A0A0Q9W9R8 A0A1A9XZQ4 A0A1B0BLC9 A0A084WG00 B4KHK8 A0A0M4EAX3 A0A1B0A005 A0A1A9V1F2 A0A336KWB3 A0A336N553 A0A182H2A3 A0A0Q9X4W1 A0A158NPL0 E9IDZ5 A0A195ETC0 A0A195DRH3 A0A151IIN2 A0A1B0D181 A0A026W1D1 A0A0M8ZVR1 A0A151X275 A0A154PQG6 A0A0L7QP79 A0A3L8DF44 A0A2M4BS19 A0A2M4BS12
Pubmed
19121390
22118469
26354079
28756777
23537049
18362917
+ More
19820115 24845553 29403074 28004739 12364791 20966253 25244985 20920257 23761445 24495485 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 22936249 15632085 25830018 19859648 25315136 17510324 24438588 26483478 18057021 21347285 21282665 24508170 30249741
19820115 24845553 29403074 28004739 12364791 20966253 25244985 20920257 23761445 24495485 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25348373 22936249 15632085 25830018 19859648 25315136 17510324 24438588 26483478 18057021 21347285 21282665 24508170 30249741
EMBL
BABH01001834
BABH01001835
ODYU01000548
SOQ35495.1
AGBW02013786
OWR42819.1
+ More
NWSH01002819 PCG67483.1 KQ459604 KPI92612.1 KZ150030 PZC74755.1 KQ461196 KPJ06563.1 JN033720 AEW46873.1 APGK01035175 KB740923 ENN78214.1 KB632326 ERL92495.1 KQ971354 KYB26332.1 KK852636 KDR19715.1 GEDC01010582 JAS26716.1 GEDC01026939 JAS10359.1 PYGN01000159 PSN52523.1 GEZM01054507 JAV73601.1 AAAB01008984 GEZM01054505 JAV73602.1 EAA14908.4 CVRI01000067 CRL06840.1 CVRI01000058 CRL02564.1 AXCM01000998 AJWK01010180 AJWK01034916 ADMH02001595 ETN61927.1 APCN01004232 GGFL01003390 MBW67568.1 GEZM01054508 JAV73600.1 GAMC01013402 JAB93153.1 CH963920 EDW77735.2 AE014134 AY089692 AAG22440.2 AAL90430.1 CM000361 EDX05467.1 CM000158 EDW90373.1 CH480822 EDW55716.1 GAKP01023242 JAC35714.1 GDHF01017111 JAI35203.1 CM002910 KMY90938.1 CH902620 EDV31139.1 CH379061 KRT04933.1 GBXI01011922 JAD02370.1 FJ821025 ACN94619.1 EAL33350.1 GAMC01013403 JAB93152.1 CH479195 EDW27244.1 OUUW01000010 SPP85532.1 CH954179 EDV54697.2 CH477353 EAT42848.1 KK856790 PTY26418.1 CH940654 KRF77957.1 EDW57791.1 CH916368 EDW03159.1 KRF77956.1 JXJN01016283 ATLV01023416 KE525343 KFB49144.1 CH933807 EDW12287.1 CP012523 ALC38460.1 UFQS01001105 UFQT01001105 SSX09027.1 SSX28938.1 UFQT01002506 SSX33598.1 JXUM01104931 JXUM01104932 JXUM01104933 JXUM01104934 KQ564927 KXJ71565.1 KRG03174.1 KRG03175.1 ADTU01001597 GL762556 EFZ21143.1 KQ981979 KYN31406.1 KQ980612 KYN15104.1 KQ977474 KYN02511.1 AJVK01002440 KK107519 EZA49391.1 KQ435859 KOX70593.1 KQ982585 KYQ54356.1 KQ435007 KZC13498.1 KQ414842 KOC60359.1 QOIP01000009 RLU19090.1 GGFJ01006671 MBW55812.1 GGFJ01006726 MBW55867.1
NWSH01002819 PCG67483.1 KQ459604 KPI92612.1 KZ150030 PZC74755.1 KQ461196 KPJ06563.1 JN033720 AEW46873.1 APGK01035175 KB740923 ENN78214.1 KB632326 ERL92495.1 KQ971354 KYB26332.1 KK852636 KDR19715.1 GEDC01010582 JAS26716.1 GEDC01026939 JAS10359.1 PYGN01000159 PSN52523.1 GEZM01054507 JAV73601.1 AAAB01008984 GEZM01054505 JAV73602.1 EAA14908.4 CVRI01000067 CRL06840.1 CVRI01000058 CRL02564.1 AXCM01000998 AJWK01010180 AJWK01034916 ADMH02001595 ETN61927.1 APCN01004232 GGFL01003390 MBW67568.1 GEZM01054508 JAV73600.1 GAMC01013402 JAB93153.1 CH963920 EDW77735.2 AE014134 AY089692 AAG22440.2 AAL90430.1 CM000361 EDX05467.1 CM000158 EDW90373.1 CH480822 EDW55716.1 GAKP01023242 JAC35714.1 GDHF01017111 JAI35203.1 CM002910 KMY90938.1 CH902620 EDV31139.1 CH379061 KRT04933.1 GBXI01011922 JAD02370.1 FJ821025 ACN94619.1 EAL33350.1 GAMC01013403 JAB93152.1 CH479195 EDW27244.1 OUUW01000010 SPP85532.1 CH954179 EDV54697.2 CH477353 EAT42848.1 KK856790 PTY26418.1 CH940654 KRF77957.1 EDW57791.1 CH916368 EDW03159.1 KRF77956.1 JXJN01016283 ATLV01023416 KE525343 KFB49144.1 CH933807 EDW12287.1 CP012523 ALC38460.1 UFQS01001105 UFQT01001105 SSX09027.1 SSX28938.1 UFQT01002506 SSX33598.1 JXUM01104931 JXUM01104932 JXUM01104933 JXUM01104934 KQ564927 KXJ71565.1 KRG03174.1 KRG03175.1 ADTU01001597 GL762556 EFZ21143.1 KQ981979 KYN31406.1 KQ980612 KYN15104.1 KQ977474 KYN02511.1 AJVK01002440 KK107519 EZA49391.1 KQ435859 KOX70593.1 KQ982585 KYQ54356.1 KQ435007 KZC13498.1 KQ414842 KOC60359.1 QOIP01000009 RLU19090.1 GGFJ01006671 MBW55812.1 GGFJ01006726 MBW55867.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000019118
+ More
UP000030742 UP000007266 UP000027135 UP000192223 UP000245037 UP000076407 UP000075903 UP000075882 UP000075881 UP000007062 UP000076408 UP000075902 UP000183832 UP000075880 UP000075884 UP000075883 UP000092461 UP000000673 UP000075900 UP000075840 UP000069272 UP000007798 UP000000803 UP000000304 UP000002282 UP000001292 UP000007801 UP000001819 UP000192221 UP000008744 UP000095300 UP000268350 UP000095301 UP000008711 UP000008820 UP000075885 UP000075920 UP000091820 UP000008792 UP000001070 UP000092443 UP000092460 UP000030765 UP000009192 UP000092553 UP000092445 UP000078200 UP000069940 UP000249989 UP000005205 UP000078541 UP000078492 UP000078542 UP000092462 UP000053097 UP000053105 UP000075809 UP000076502 UP000053825 UP000279307
UP000030742 UP000007266 UP000027135 UP000192223 UP000245037 UP000076407 UP000075903 UP000075882 UP000075881 UP000007062 UP000076408 UP000075902 UP000183832 UP000075880 UP000075884 UP000075883 UP000092461 UP000000673 UP000075900 UP000075840 UP000069272 UP000007798 UP000000803 UP000000304 UP000002282 UP000001292 UP000007801 UP000001819 UP000192221 UP000008744 UP000095300 UP000268350 UP000095301 UP000008711 UP000008820 UP000075885 UP000075920 UP000091820 UP000008792 UP000001070 UP000092443 UP000092460 UP000030765 UP000009192 UP000092553 UP000092445 UP000078200 UP000069940 UP000249989 UP000005205 UP000078541 UP000078492 UP000078542 UP000092462 UP000053097 UP000053105 UP000075809 UP000076502 UP000053825 UP000279307
Interpro
IPR001254
Trypsin_dom
+ More
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR038565 CLIP_sf
IPR022700 CLIP
IPR001320 Iontro_rcpt
IPR004821 Cyt_trans-like
IPR041723 CCT
IPR014729 Rossmann-like_a/b/a_fold
IPR017946 PLC-like_Pdiesterase_TIM-brl
IPR000909 PLipase_C_PInositol-sp_X_dom
IPR033116 TRYPSIN_SER
IPR018114 TRYPSIN_HIS
IPR009003 Peptidase_S1_PA
IPR001314 Peptidase_S1A
IPR038565 CLIP_sf
IPR022700 CLIP
IPR001320 Iontro_rcpt
IPR004821 Cyt_trans-like
IPR041723 CCT
IPR014729 Rossmann-like_a/b/a_fold
IPR017946 PLC-like_Pdiesterase_TIM-brl
IPR000909 PLipase_C_PInositol-sp_X_dom
IPR033116 TRYPSIN_SER
Gene 3D
ProteinModelPortal
H9JG24
A0A2H1V3S7
A0A212EMW9
A0A2A4J7W5
A0A194PH36
A0A2W1BMA8
+ More
A0A194QLY6 G9F9H5 N6TD06 U4UR11 A0A139WEJ1 A0A067RAE5 A0A1W4X8T0 A0A1B6DLX7 A0A1B6CAJ6 A0A2P8Z7P1 A0A1Y1LL09 A0A182WW36 A0A182UVC0 A0A1S4H1C9 A0A182L6D4 A0A182KB17 A0A1Y1LIX7 Q7PVJ0 A0A182YDQ8 A0A182U9R2 A0A1J1J398 A0A1J1ISM1 A0A182JCH5 A0A182NU45 A0A182M604 A0A1B0GLG1 W5JC77 A0A182RIP5 A0A182HGH5 A0A2M4CRS5 A0A1Y1LML9 W8B7Y4 A0A3F2YQ98 B4N0X2 Q8SXE1 B4Q9G0 B4PAG5 B4I5R0 A0A034UZP8 A0A0K8V8U7 A0A0J9R4E3 B3MNQ2 A0A0R3P0F0 A0A0A1WV38 D0QWC2 A0A1W4W8K3 Q29K28 W8B3K9 B4GXD3 A0A1I8PM58 A0A3B0JYM3 A0A1I8MF00 B3NM97 Q179F8 A0A182PE86 A0A1S4FB72 A0A2R7X3P5 A0A182VUG5 A0A1A9W0P4 A0A0Q9WBS0 B4M9B5 B4JCK5 A0A0Q9W9R8 A0A1A9XZQ4 A0A1B0BLC9 A0A084WG00 B4KHK8 A0A0M4EAX3 A0A1B0A005 A0A1A9V1F2 A0A336KWB3 A0A336N553 A0A182H2A3 A0A0Q9X4W1 A0A158NPL0 E9IDZ5 A0A195ETC0 A0A195DRH3 A0A151IIN2 A0A1B0D181 A0A026W1D1 A0A0M8ZVR1 A0A151X275 A0A154PQG6 A0A0L7QP79 A0A3L8DF44 A0A2M4BS19 A0A2M4BS12
A0A194QLY6 G9F9H5 N6TD06 U4UR11 A0A139WEJ1 A0A067RAE5 A0A1W4X8T0 A0A1B6DLX7 A0A1B6CAJ6 A0A2P8Z7P1 A0A1Y1LL09 A0A182WW36 A0A182UVC0 A0A1S4H1C9 A0A182L6D4 A0A182KB17 A0A1Y1LIX7 Q7PVJ0 A0A182YDQ8 A0A182U9R2 A0A1J1J398 A0A1J1ISM1 A0A182JCH5 A0A182NU45 A0A182M604 A0A1B0GLG1 W5JC77 A0A182RIP5 A0A182HGH5 A0A2M4CRS5 A0A1Y1LML9 W8B7Y4 A0A3F2YQ98 B4N0X2 Q8SXE1 B4Q9G0 B4PAG5 B4I5R0 A0A034UZP8 A0A0K8V8U7 A0A0J9R4E3 B3MNQ2 A0A0R3P0F0 A0A0A1WV38 D0QWC2 A0A1W4W8K3 Q29K28 W8B3K9 B4GXD3 A0A1I8PM58 A0A3B0JYM3 A0A1I8MF00 B3NM97 Q179F8 A0A182PE86 A0A1S4FB72 A0A2R7X3P5 A0A182VUG5 A0A1A9W0P4 A0A0Q9WBS0 B4M9B5 B4JCK5 A0A0Q9W9R8 A0A1A9XZQ4 A0A1B0BLC9 A0A084WG00 B4KHK8 A0A0M4EAX3 A0A1B0A005 A0A1A9V1F2 A0A336KWB3 A0A336N553 A0A182H2A3 A0A0Q9X4W1 A0A158NPL0 E9IDZ5 A0A195ETC0 A0A195DRH3 A0A151IIN2 A0A1B0D181 A0A026W1D1 A0A0M8ZVR1 A0A151X275 A0A154PQG6 A0A0L7QP79 A0A3L8DF44 A0A2M4BS19 A0A2M4BS12
PDB
2OLG
E-value=6.08828e-40,
Score=412
Ontologies
GO
Topology
SignalP
Position: 1 - 20,
Likelihood: 0.987467
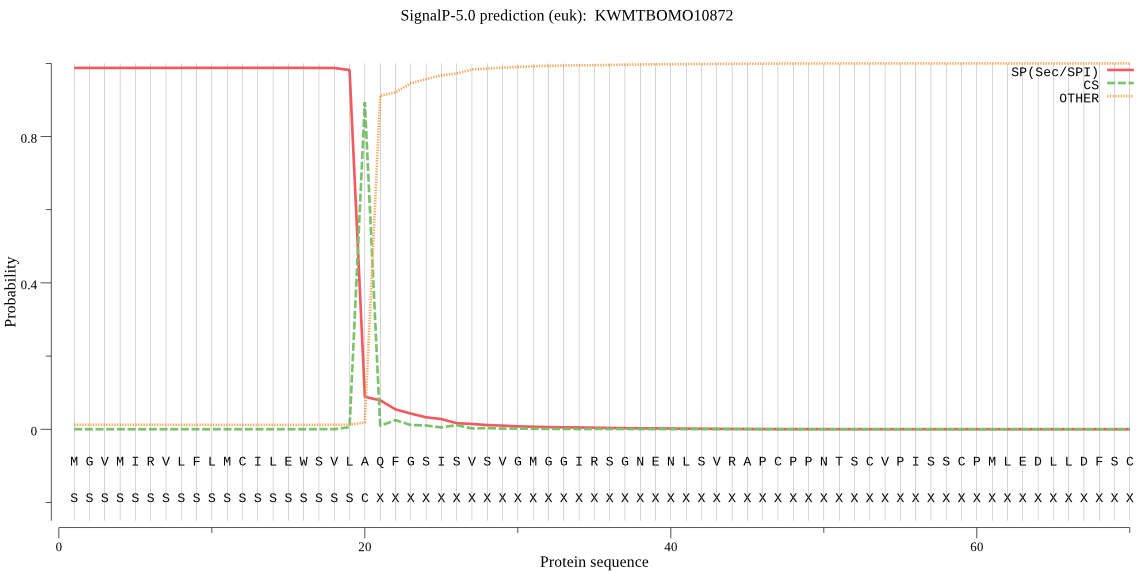
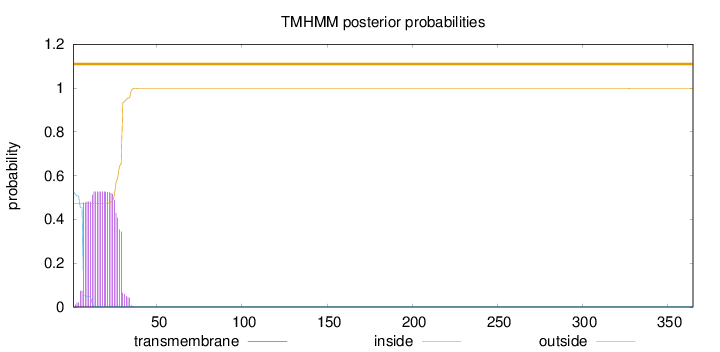
Length:
365
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
11.69864
Exp number, first 60 AAs:
11.69758
Total prob of N-in:
0.52679
POSSIBLE N-term signal
sequence
outside
1 - 365
Population Genetic Test Statistics
Pi
237.061159
Theta
136.627391
Tajima's D
1.789291
CLR
0.497515
CSRT
0.839858007099645
Interpretation
Uncertain
