Gene
KWMTBOMO10868
Annotation
PREDICTED:_cilia-_and_flagella-associated_protein_61_[Bombyx_mori]
Full name
Cilia- and flagella-associated protein 61
Location in the cell
PlasmaMembrane Reliability : 3.165
Sequence
CDS
ATGGTTTTATCTTATTCAGTTTTCGAACCACATTTCCGATTTTTCGCAAGAGATATCATGCGATTGTCTGGAGCCAGTGCCTTGATTTGGTTGACGGGTTACAGAAATAAATGGGTGATGAAGAAAGCCAATTCATTAGCGTCAGTGATGATTCCATTGATGCCTTTAGAGTCAGCCGTAGACTACGTTATACCAGATTTCAAAAGTGATTTAACGAATAAAACATTCTCGTTTAGCGCTTGGTTTTTGCCGAAAAAACTGACCAGTGTACCAATGTTGTACGTAACAACACGTATAGTAGTCGTGGGTGCTTCAAAAACTGCAATGTCCTTTCTGAATTCTTTATTATTTAGGTAA
Protein
MVLSYSVFEPHFRFFARDIMRLSGASALIWLTGYRNKWVMKKANSLASVMIPLMPLESAVDYVIPDFKSDLTNKTFSFSAWFLPKKLTSVPMLYVTTRIVVVGASKTAMSFLNSLLFR
Summary
Description
May regulate cilium motility through its role in the assembly of the axonemal radial spokes.
Keywords
Alternative splicing
Cell projection
Complete proteome
Cytoplasm
Cytoskeleton
Reference proteome
Polymorphism
Feature
chain Cilia- and flagella-associated protein 61
splice variant In isoform 2.
sequence variant In dbSNP:rs17852602.
splice variant In isoform 2.
sequence variant In dbSNP:rs17852602.
Uniprot
A0A194QNL7
A0A212EMU9
A0A194PHU0
A0A2W1BIR6
A0A2A4J7R5
A0A0L7LJB0
+ More
A0A2H1WN36 A0A194QSS1 A0A2A4K947 A0A212ESY4 A0A2W1BGT5 A0A212EW30 G1PUU8 E0W2I6 A0A1U7R8Q0 A0A2U4AQR4 A0A2Y9SU25 A0A341C6A3 I3MNK3 A0A3Q0CU67 A0A3N0Z1T7 A0A384AX21 A0A340WZU5 A0A1B6EM11 L5JZL6 G3GVV1 F7HC74 L8YF39 A0A2Y9PSU3 A0A1B6FN19 A0A2K5QPV4 W5NX33 F6X556 A0A151NDD8 Q0P4Q2 A0A2K6UFJ5 S4RNJ1 M3VVA1 F7EI69 A0A2I2UGX1 A0A2Y9IRX1 F6RHF7 A0A2K6GKD5 A0A1S3G117 M3XQ59 A0A1B6EFB2 G1T9L3 A8Y5L4 A0A1B6DDD5 A0A1B6DB85 A0A1V4IH35 A0A1V4IGL0 B3RKX7 L8IZ84 Q8CEL2 E2A7S6 A0A1B6E7N2 A0A1B6DFX1 A0A2U3ZTI5 A0A2U3Y1T8 A0A195FK74 A0A3Q7PFT7 A0A3Q7PG93 A0A0D9RFU5 A0A1L8G7Z5 A0A3Q7SCW0 E2BP73 H0X516 E1BP40 E2RFZ5 A0A2K5IST5 A0A3L8DJ53 A0A026WTI8 A0A2B4SFT9 G3WL91 A0A2K5ZVJ7 A0A2K6JV94 A0A2K6CTS3 A0A2K6QX22 A0A2K5NX28 A0A2K5TNN9 H0Y3Y7 G7PH29 G7N329 A0A3B1KA51 W5LGB0 B7Z8Z0 A0A2J8VDG8 A0A2K6JVA0 Q8NHU2 A0A337SDA7 A0A096NVQ3 A0A2K5TNN6 Q8NHU2-4 H2QK15 A0A369SDN4 A0A2J8VDH1
A0A2H1WN36 A0A194QSS1 A0A2A4K947 A0A212ESY4 A0A2W1BGT5 A0A212EW30 G1PUU8 E0W2I6 A0A1U7R8Q0 A0A2U4AQR4 A0A2Y9SU25 A0A341C6A3 I3MNK3 A0A3Q0CU67 A0A3N0Z1T7 A0A384AX21 A0A340WZU5 A0A1B6EM11 L5JZL6 G3GVV1 F7HC74 L8YF39 A0A2Y9PSU3 A0A1B6FN19 A0A2K5QPV4 W5NX33 F6X556 A0A151NDD8 Q0P4Q2 A0A2K6UFJ5 S4RNJ1 M3VVA1 F7EI69 A0A2I2UGX1 A0A2Y9IRX1 F6RHF7 A0A2K6GKD5 A0A1S3G117 M3XQ59 A0A1B6EFB2 G1T9L3 A8Y5L4 A0A1B6DDD5 A0A1B6DB85 A0A1V4IH35 A0A1V4IGL0 B3RKX7 L8IZ84 Q8CEL2 E2A7S6 A0A1B6E7N2 A0A1B6DFX1 A0A2U3ZTI5 A0A2U3Y1T8 A0A195FK74 A0A3Q7PFT7 A0A3Q7PG93 A0A0D9RFU5 A0A1L8G7Z5 A0A3Q7SCW0 E2BP73 H0X516 E1BP40 E2RFZ5 A0A2K5IST5 A0A3L8DJ53 A0A026WTI8 A0A2B4SFT9 G3WL91 A0A2K5ZVJ7 A0A2K6JV94 A0A2K6CTS3 A0A2K6QX22 A0A2K5NX28 A0A2K5TNN9 H0Y3Y7 G7PH29 G7N329 A0A3B1KA51 W5LGB0 B7Z8Z0 A0A2J8VDG8 A0A2K6JVA0 Q8NHU2 A0A337SDA7 A0A096NVQ3 A0A2K5TNN6 Q8NHU2-4 H2QK15 A0A369SDN4 A0A2J8VDH1
Pubmed
26354079
22118469
28756777
26227816
21993624
20566863
+ More
26319212 23258410 21804562 25243066 23385571 20809919 20431018 22293439 17975172 19892987 19468303 21183079 18719581 22751099 15489334 16141072 20798317 27762356 19393038 16341006 30249741 24508170 21709235 25362486 11780052 22002653 25329095 14702039 17974005 23033978 16136131 30042472
26319212 23258410 21804562 25243066 23385571 20809919 20431018 22293439 17975172 19892987 19468303 21183079 18719581 22751099 15489334 16141072 20798317 27762356 19393038 16341006 30249741 24508170 21709235 25362486 11780052 22002653 25329095 14702039 17974005 23033978 16136131 30042472
EMBL
KQ461196
KPJ06565.1
AGBW02013786
OWR42822.1
KQ459604
KPI92613.1
+ More
KZ150030 PZC74758.1 NWSH01002819 PCG67480.1 JTDY01000961 KOB75271.1 ODYU01009812 SOQ54479.1 KPJ06586.1 NWSH01000028 PCG80579.1 AGBW02012686 OWR44606.1 KZ150058 PZC74272.1 AGBW02012095 OWR45689.1 AAPE02034453 AAPE02034454 AAPE02034455 DS235878 EEB19842.1 AGTP01017380 AGTP01017381 AGTP01017382 AGTP01017383 RJVU01015592 ROL52420.1 GECZ01030837 JAS38932.1 KB031072 ELK03926.1 JH000044 EGV95388.1 GAMS01006521 JAB16615.1 KB363509 ELV12996.1 GECZ01018164 JAS51605.1 AMGL01024972 AMGL01024973 AMGL01024974 AMGL01024975 AMGL01024976 AMGL01024977 AMGL01024978 AMGL01024979 AMGL01024980 AMGL01024981 AAMC01117902 AAMC01117903 AAMC01117904 AAMC01117905 AAMC01117906 AAMC01117907 AAMC01117908 AKHW03003332 KYO34750.1 BC121953 AAI21954.1 AANG04003379 AEYP01035767 AEYP01035768 AEYP01035769 AEYP01035770 AEYP01035771 GEDC01000689 JAS36609.1 AAGW02052417 AAGW02052418 AAGW02052419 AAGW02052420 AAGW02052421 AL672100 AL732467 AL935056 GEDC01013609 JAS23689.1 GEDC01014370 JAS22928.1 LSYS01009753 OPJ58985.1 OPJ58984.1 DS985241 EDV28663.1 JH880468 ELR61243.1 BC024760 BC107030 BC107031 AK019712 GL437384 EFN70506.1 GEDC01003347 JAS33951.1 GEDC01012714 JAS24584.1 KQ981512 KYN40803.1 AQIB01145295 AQIB01145296 AQIB01145297 AQIB01145298 CM004474 OCT79814.1 GL449553 EFN82493.1 AAQR03145489 AAQR03145490 AAQR03145491 AAQR03145492 AAQR03145493 AAQR03145494 AAQR03145495 AAQR03145496 AAQR03145497 AAQR03145498 AAEX03013754 QOIP01000008 RLU19878.1 KK107109 EZA59327.1 LSMT01000103 PFX27427.1 AEFK01029396 AEFK01029397 AEFK01029398 AEFK01029399 AEFK01029400 AEFK01029401 AEFK01029402 AEFK01029403 AEFK01029404 AEFK01029405 AQIA01008002 AQIA01008003 AL035454 AL049648 AL121721 AL158849 AL161658 KF456848 CM001285 EHH65585.1 CM001262 EHH19961.1 AK304174 BAH14126.1 NDHI03003422 PNJ55561.1 AK057364 AL117439 BC028708 BC031674 AHZZ02000703 AHZZ02000704 AHZZ02000705 AHZZ02000706 AACZ04064887 AACZ04064888 AACZ04064889 AACZ04064890 AACZ04064891 AACZ04064892 AACZ04064893 NBAG03000331 PNI39501.1 NOWV01000020 RDD45035.1 PNJ55569.1
KZ150030 PZC74758.1 NWSH01002819 PCG67480.1 JTDY01000961 KOB75271.1 ODYU01009812 SOQ54479.1 KPJ06586.1 NWSH01000028 PCG80579.1 AGBW02012686 OWR44606.1 KZ150058 PZC74272.1 AGBW02012095 OWR45689.1 AAPE02034453 AAPE02034454 AAPE02034455 DS235878 EEB19842.1 AGTP01017380 AGTP01017381 AGTP01017382 AGTP01017383 RJVU01015592 ROL52420.1 GECZ01030837 JAS38932.1 KB031072 ELK03926.1 JH000044 EGV95388.1 GAMS01006521 JAB16615.1 KB363509 ELV12996.1 GECZ01018164 JAS51605.1 AMGL01024972 AMGL01024973 AMGL01024974 AMGL01024975 AMGL01024976 AMGL01024977 AMGL01024978 AMGL01024979 AMGL01024980 AMGL01024981 AAMC01117902 AAMC01117903 AAMC01117904 AAMC01117905 AAMC01117906 AAMC01117907 AAMC01117908 AKHW03003332 KYO34750.1 BC121953 AAI21954.1 AANG04003379 AEYP01035767 AEYP01035768 AEYP01035769 AEYP01035770 AEYP01035771 GEDC01000689 JAS36609.1 AAGW02052417 AAGW02052418 AAGW02052419 AAGW02052420 AAGW02052421 AL672100 AL732467 AL935056 GEDC01013609 JAS23689.1 GEDC01014370 JAS22928.1 LSYS01009753 OPJ58985.1 OPJ58984.1 DS985241 EDV28663.1 JH880468 ELR61243.1 BC024760 BC107030 BC107031 AK019712 GL437384 EFN70506.1 GEDC01003347 JAS33951.1 GEDC01012714 JAS24584.1 KQ981512 KYN40803.1 AQIB01145295 AQIB01145296 AQIB01145297 AQIB01145298 CM004474 OCT79814.1 GL449553 EFN82493.1 AAQR03145489 AAQR03145490 AAQR03145491 AAQR03145492 AAQR03145493 AAQR03145494 AAQR03145495 AAQR03145496 AAQR03145497 AAQR03145498 AAEX03013754 QOIP01000008 RLU19878.1 KK107109 EZA59327.1 LSMT01000103 PFX27427.1 AEFK01029396 AEFK01029397 AEFK01029398 AEFK01029399 AEFK01029400 AEFK01029401 AEFK01029402 AEFK01029403 AEFK01029404 AEFK01029405 AQIA01008002 AQIA01008003 AL035454 AL049648 AL121721 AL158849 AL161658 KF456848 CM001285 EHH65585.1 CM001262 EHH19961.1 AK304174 BAH14126.1 NDHI03003422 PNJ55561.1 AK057364 AL117439 BC028708 BC031674 AHZZ02000703 AHZZ02000704 AHZZ02000705 AHZZ02000706 AACZ04064887 AACZ04064888 AACZ04064889 AACZ04064890 AACZ04064891 AACZ04064892 AACZ04064893 NBAG03000331 PNI39501.1 NOWV01000020 RDD45035.1 PNJ55569.1
Proteomes
UP000053240
UP000007151
UP000053268
UP000218220
UP000037510
UP000001074
+ More
UP000009046 UP000189705 UP000245320 UP000252040 UP000005215 UP000189706 UP000261681 UP000265300 UP000010552 UP000001075 UP000008225 UP000011518 UP000248483 UP000233040 UP000002356 UP000008143 UP000050525 UP000233220 UP000245300 UP000011712 UP000248482 UP000002281 UP000233160 UP000081671 UP000000715 UP000001811 UP000000589 UP000190648 UP000009022 UP000000311 UP000245340 UP000245341 UP000078541 UP000286641 UP000029965 UP000186698 UP000286640 UP000008237 UP000005225 UP000009136 UP000002254 UP000233080 UP000279307 UP000053097 UP000225706 UP000007648 UP000233140 UP000233180 UP000233120 UP000233200 UP000233060 UP000233100 UP000005640 UP000009130 UP000018467 UP000028761 UP000002277 UP000253843
UP000009046 UP000189705 UP000245320 UP000252040 UP000005215 UP000189706 UP000261681 UP000265300 UP000010552 UP000001075 UP000008225 UP000011518 UP000248483 UP000233040 UP000002356 UP000008143 UP000050525 UP000233220 UP000245300 UP000011712 UP000248482 UP000002281 UP000233160 UP000081671 UP000000715 UP000001811 UP000000589 UP000190648 UP000009022 UP000000311 UP000245340 UP000245341 UP000078541 UP000286641 UP000029965 UP000186698 UP000286640 UP000008237 UP000005225 UP000009136 UP000002254 UP000233080 UP000279307 UP000053097 UP000225706 UP000007648 UP000233140 UP000233180 UP000233120 UP000233200 UP000233060 UP000233100 UP000005640 UP000009130 UP000018467 UP000028761 UP000002277 UP000253843
Interpro
IPR032151
CFAP61_N
+ More
IPR036188 FAD/NAD-bd_sf
IPR038884 CFAP61
IPR001005 SANT/Myb
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR000949 ELM2_dom
IPR016181 Acyl_CoA_acyltransferase
IPR023753 FAD/NAD-binding_dom
IPR032675 LRR_dom_sf
IPR016024 ARM-type_fold
IPR029058 AB_hydrolase
IPR001356 Homeobox_dom
IPR024983 CHAT_dom
IPR011990 TPR-like_helical_dom_sf
IPR036188 FAD/NAD-bd_sf
IPR038884 CFAP61
IPR001005 SANT/Myb
IPR017884 SANT_dom
IPR009057 Homeobox-like_sf
IPR000949 ELM2_dom
IPR016181 Acyl_CoA_acyltransferase
IPR023753 FAD/NAD-binding_dom
IPR032675 LRR_dom_sf
IPR016024 ARM-type_fold
IPR029058 AB_hydrolase
IPR001356 Homeobox_dom
IPR024983 CHAT_dom
IPR011990 TPR-like_helical_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A194QNL7
A0A212EMU9
A0A194PHU0
A0A2W1BIR6
A0A2A4J7R5
A0A0L7LJB0
+ More
A0A2H1WN36 A0A194QSS1 A0A2A4K947 A0A212ESY4 A0A2W1BGT5 A0A212EW30 G1PUU8 E0W2I6 A0A1U7R8Q0 A0A2U4AQR4 A0A2Y9SU25 A0A341C6A3 I3MNK3 A0A3Q0CU67 A0A3N0Z1T7 A0A384AX21 A0A340WZU5 A0A1B6EM11 L5JZL6 G3GVV1 F7HC74 L8YF39 A0A2Y9PSU3 A0A1B6FN19 A0A2K5QPV4 W5NX33 F6X556 A0A151NDD8 Q0P4Q2 A0A2K6UFJ5 S4RNJ1 M3VVA1 F7EI69 A0A2I2UGX1 A0A2Y9IRX1 F6RHF7 A0A2K6GKD5 A0A1S3G117 M3XQ59 A0A1B6EFB2 G1T9L3 A8Y5L4 A0A1B6DDD5 A0A1B6DB85 A0A1V4IH35 A0A1V4IGL0 B3RKX7 L8IZ84 Q8CEL2 E2A7S6 A0A1B6E7N2 A0A1B6DFX1 A0A2U3ZTI5 A0A2U3Y1T8 A0A195FK74 A0A3Q7PFT7 A0A3Q7PG93 A0A0D9RFU5 A0A1L8G7Z5 A0A3Q7SCW0 E2BP73 H0X516 E1BP40 E2RFZ5 A0A2K5IST5 A0A3L8DJ53 A0A026WTI8 A0A2B4SFT9 G3WL91 A0A2K5ZVJ7 A0A2K6JV94 A0A2K6CTS3 A0A2K6QX22 A0A2K5NX28 A0A2K5TNN9 H0Y3Y7 G7PH29 G7N329 A0A3B1KA51 W5LGB0 B7Z8Z0 A0A2J8VDG8 A0A2K6JVA0 Q8NHU2 A0A337SDA7 A0A096NVQ3 A0A2K5TNN6 Q8NHU2-4 H2QK15 A0A369SDN4 A0A2J8VDH1
A0A2H1WN36 A0A194QSS1 A0A2A4K947 A0A212ESY4 A0A2W1BGT5 A0A212EW30 G1PUU8 E0W2I6 A0A1U7R8Q0 A0A2U4AQR4 A0A2Y9SU25 A0A341C6A3 I3MNK3 A0A3Q0CU67 A0A3N0Z1T7 A0A384AX21 A0A340WZU5 A0A1B6EM11 L5JZL6 G3GVV1 F7HC74 L8YF39 A0A2Y9PSU3 A0A1B6FN19 A0A2K5QPV4 W5NX33 F6X556 A0A151NDD8 Q0P4Q2 A0A2K6UFJ5 S4RNJ1 M3VVA1 F7EI69 A0A2I2UGX1 A0A2Y9IRX1 F6RHF7 A0A2K6GKD5 A0A1S3G117 M3XQ59 A0A1B6EFB2 G1T9L3 A8Y5L4 A0A1B6DDD5 A0A1B6DB85 A0A1V4IH35 A0A1V4IGL0 B3RKX7 L8IZ84 Q8CEL2 E2A7S6 A0A1B6E7N2 A0A1B6DFX1 A0A2U3ZTI5 A0A2U3Y1T8 A0A195FK74 A0A3Q7PFT7 A0A3Q7PG93 A0A0D9RFU5 A0A1L8G7Z5 A0A3Q7SCW0 E2BP73 H0X516 E1BP40 E2RFZ5 A0A2K5IST5 A0A3L8DJ53 A0A026WTI8 A0A2B4SFT9 G3WL91 A0A2K5ZVJ7 A0A2K6JV94 A0A2K6CTS3 A0A2K6QX22 A0A2K5NX28 A0A2K5TNN9 H0Y3Y7 G7PH29 G7N329 A0A3B1KA51 W5LGB0 B7Z8Z0 A0A2J8VDG8 A0A2K6JVA0 Q8NHU2 A0A337SDA7 A0A096NVQ3 A0A2K5TNN6 Q8NHU2-4 H2QK15 A0A369SDN4 A0A2J8VDH1
Ontologies
PANTHER
Topology
Subcellular location
Nucleus
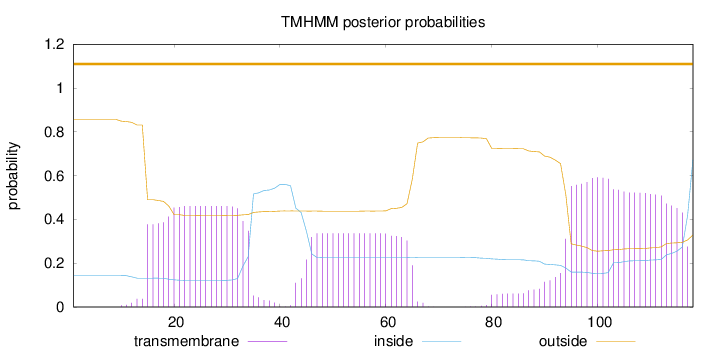
Length:
118
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
29.37096
Exp number, first 60 AAs:
14.42936
Total prob of N-in:
0.14316
POSSIBLE N-term signal
sequence
outside
1 - 118
Population Genetic Test Statistics
Pi
166.325253
Theta
109.899364
Tajima's D
1.651226
CLR
0.402213
CSRT
0.820458977051147
Interpretation
Uncertain
