Gene
KWMTBOMO10866
Pre Gene Modal
BGIBMGA008467
Annotation
PREDICTED:_protein_regulator_of_cytokinesis_1-like_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.649
Sequence
CDS
ATGATTACATTTACAGAAGACGTGAACAAGTTGTTCAATGAAGTCCTACAGGAGATTTCTAATAACATCCGTGAAATGATGGAAGAGCTCTGGTTAAACTGGTCCCATTTGGGTGTTGATGATAGCACCAAAATAAATAACATAACAAAACTCGTTCAAATCGAGAAAGAACTCCACCGAGATGTGATCTCAGAGACCAGACAAAAATTAAAAATCATGCAGGGACAAGTCGATAAACTTAAAGAGGAAACTGAAGAACTTAGCAGATGTTTATCAGTGGACATCACCATATTGGATTTCAAAGAGGAAATGATGCTCGTTGAATATAAGAGAGAGTTAGAGGAACAAATCTCTGGTTATAGAGAACAAGTTGAACAACGTCGTATGAAGATGGAACGGCTTTTAGAATGGCAGAGAGACCTTGTAGACAAGCTGGGCGTTACCATCCATGAGCTTCAAGAAATACCACTCCCCTCTGAAGAAGAGCTTGATAGATTAAAGGATCATCTGGAGAAATTGCAAGCTGAAAGAGATAAGAGGGCAGAGATATTCTTAAATGTTCAAGTTGAGGTCAAAGATATTATGGAAAAACTACAAATAAAGCCACATAATAAGTTTGAACATATTGTAACTTCATCTTTATCAGTGGATTTTAAAGTAACAGATCTAAACATGGATCGCCTCGCCAAACTTCAACAAGATTTGCAGGAGAAATTTGAGCAGACTAATAACAGGGTGCTGGAGCTTCGTGAAAAATTATCCAAGTTATGGGATTGTCTTGATGAAGATCAGATATACAGAGAGAACTTCCTTCATGCCCATCCTGGCTGTCACCCTCAGACTGAATCTGCTATCAAAGAGGAGATCAAGAGATGCGAACAAATCAAACGGCAGAAAATTCAGGTGTTTGTGGCGAATGTACGTACGAAGATCAAGTTGATGTGGGACAATGTGATGTACTCGACCGATGAGCGTGAGGAGTTTGTGCATTACTACCAGGATATTTTCACTGAGGACACCCTCACTCTACACGAATTATATCTTGAAAAGATAACTAAATTTTACAATGAAAATAAGAATATATTTGATTTGGTGCTGACACGCAAGAATCTGTGGCTGAAGATGACAGAATTAGAGGCGAGAGCTTCCGAGCCTGGCCGCTACCACAACCGGGGTGGACAACTGCTACGAGAAGAGAAAGAAAGAAAAGCTATTGCTAGCAATCTTCCCAAAATAGAAGCACAGTTAAGGGAATTGGTATCTGAGTACGAAGCCAAGACAGGATCTACCTTTACTGTTGATGGTAAACCCCTGTTGCAACTAATGGAAGACGAGTGGGAAGCTCGAAAGGCGGAGCGCCACAACAAGCTGTCGGCCCGCAAGCAGGCGCTGACGCCCACCACGCCGCTGCTGCGCTCGCTGGCCACGTCGCCGCTCGGCAAGCGGAACCGCACCGCCGCCGGCCTCGCGCACACCGCCGACAGAAACAAGCCTCCGACGAAGAGGCAGCTCATCACGGGCAGCGCCACGAAAGCGGTCACCACGTTCACCAACAACCTGTCTGCCTTCAAAAGATCTGCTATTACAACTGTCAAGAGGCGTATCAGTGGTCGTTTGGCGGCCAGAGCGGTAGTCGAGGGTAAGGGTGACCATGCCAAGAGGAAGCTCGACTATGGAAACGAAGGGAAGAAAACACCTAAAGTCAACGGCAGTATTTTAAAACATAAGCGTACCTCTCATGGCAAGCGCCGGAGCAGCAGCCGCAAGTCAATGAACGCGACGCGCTCAACTGAAGACGAACGAATTTCGAACAAAGACCCGCTAATGGAGACCACGCTTCTAACCACTTACACTGATTTCAAGGACGGTATAAACGAACGTCAGATCAGTCGCAGTTCGATGGCCAATCCTAAACCGCAGGAAGGCCTCGTGCTGCCCACGATTATTTGTTCAAAGATCGAAGAGAACTCTCCGGTCACGCCAAAAAAAACTCCGAGGACGAAAACGCCACTGACCCCTAAAATAGGCAAAGAGAATATACAGCACATAATGGCGGTCACGCCCAAGAATAATCTATTGTACACACCCACGCGACTAACGCGGTCGGCTCTTAAACTGAACAATGACGGTTTCGCGACGCCCCGGGCTCCATTGAGCGCAACTAAAGTAAACCTACAGCGGCAGAACACGGTGACGGGAATGTCATTGAAAACCACCCCGAACAACGTCAGCCGATCTAAATCTCACTCGCATTTGGTCAGAGTTAAGAATCTACCGCCCCTCATCTGA
Protein
MITFTEDVNKLFNEVLQEISNNIREMMEELWLNWSHLGVDDSTKINNITKLVQIEKELHRDVISETRQKLKIMQGQVDKLKEETEELSRCLSVDITILDFKEEMMLVEYKRELEEQISGYREQVEQRRMKMERLLEWQRDLVDKLGVTIHELQEIPLPSEEELDRLKDHLEKLQAERDKRAEIFLNVQVEVKDIMEKLQIKPHNKFEHIVTSSLSVDFKVTDLNMDRLAKLQQDLQEKFEQTNNRVLELREKLSKLWDCLDEDQIYRENFLHAHPGCHPQTESAIKEEIKRCEQIKRQKIQVFVANVRTKIKLMWDNVMYSTDEREEFVHYYQDIFTEDTLTLHELYLEKITKFYNENKNIFDLVLTRKNLWLKMTELEARASEPGRYHNRGGQLLREEKERKAIASNLPKIEAQLRELVSEYEAKTGSTFTVDGKPLLQLMEDEWEARKAERHNKLSARKQALTPTTPLLRSLATSPLGKRNRTAAGLAHTADRNKPPTKRQLITGSATKAVTTFTNNLSAFKRSAITTVKRRISGRLAARAVVEGKGDHAKRKLDYGNEGKKTPKVNGSILKHKRTSHGKRRSSSRKSMNATRSTEDERISNKDPLMETTLLTTYTDFKDGINERQISRSSMANPKPQEGLVLPTIICSKIEENSPVTPKKTPRTKTPLTPKIGKENIQHIMAVTPKNNLLYTPTRLTRSALKLNNDGFATPRAPLSATKVNLQRQNTVTGMSLKTTPNNVSRSKSHSHLVRVKNLPPLI
Summary
Uniprot
H9JG20
A0A2W1BI68
A0A2H1WZH4
A0A194PNC1
A0A212EMV6
A0A195D4E0
+ More
A0A0J7KTZ3 F4W6K3 E2AJX3 E2C022 A0A195BUV6 A0A158P2A8 A0A1I8QED3 A0A151X247 A0A0K8WB47 A0A034WIK9 A0A1A9Y1R7 A0A2J7QBX1 A0A2J7QBV5 A0A2J7QBV4 A0A182ILY7 A0A182P1S9 A0A1A9ZCE6 C3XZQ0 A0A182W9E4 A0A1A9XUK9 A0A0T6BDY8 A0A336LY44 A0A1B0FIC5 A0A1B0AMG4 A0A0P4YVS4 A0A0P5G713
A0A0J7KTZ3 F4W6K3 E2AJX3 E2C022 A0A195BUV6 A0A158P2A8 A0A1I8QED3 A0A151X247 A0A0K8WB47 A0A034WIK9 A0A1A9Y1R7 A0A2J7QBX1 A0A2J7QBV5 A0A2J7QBV4 A0A182ILY7 A0A182P1S9 A0A1A9ZCE6 C3XZQ0 A0A182W9E4 A0A1A9XUK9 A0A0T6BDY8 A0A336LY44 A0A1B0FIC5 A0A1B0AMG4 A0A0P4YVS4 A0A0P5G713
EMBL
BABH01001825
KZ150030
PZC74759.1
ODYU01012224
SOQ58418.1
KQ459604
+ More
KPI92615.1 AGBW02013786 OWR42824.1 KQ976885 KYN07299.1 LBMM01003135 KMQ93917.1 GL887707 EGI70338.1 GL440100 EFN66289.1 GL451712 EFN78718.1 KQ976408 KYM91132.1 ADTU01007050 KQ982580 KYQ54503.1 GDHF01004234 JAI48080.1 GAKP01003548 GAKP01003546 GAKP01003544 GAKP01003542 JAC55406.1 NEVH01016296 PNF26068.1 PNF26070.1 PNF26066.1 GG666476 EEN66448.1 LJIG01001366 KRT85574.1 UFQT01000198 SSX21593.1 CCAG010020272 JXJN01000423 GDIP01222924 JAJ00478.1 GDIQ01245385 JAK06340.1
KPI92615.1 AGBW02013786 OWR42824.1 KQ976885 KYN07299.1 LBMM01003135 KMQ93917.1 GL887707 EGI70338.1 GL440100 EFN66289.1 GL451712 EFN78718.1 KQ976408 KYM91132.1 ADTU01007050 KQ982580 KYQ54503.1 GDHF01004234 JAI48080.1 GAKP01003548 GAKP01003546 GAKP01003544 GAKP01003542 JAC55406.1 NEVH01016296 PNF26068.1 PNF26070.1 PNF26066.1 GG666476 EEN66448.1 LJIG01001366 KRT85574.1 UFQT01000198 SSX21593.1 CCAG010020272 JXJN01000423 GDIP01222924 JAJ00478.1 GDIQ01245385 JAK06340.1
Proteomes
PRIDE
ProteinModelPortal
H9JG20
A0A2W1BI68
A0A2H1WZH4
A0A194PNC1
A0A212EMV6
A0A195D4E0
+ More
A0A0J7KTZ3 F4W6K3 E2AJX3 E2C022 A0A195BUV6 A0A158P2A8 A0A1I8QED3 A0A151X247 A0A0K8WB47 A0A034WIK9 A0A1A9Y1R7 A0A2J7QBX1 A0A2J7QBV5 A0A2J7QBV4 A0A182ILY7 A0A182P1S9 A0A1A9ZCE6 C3XZQ0 A0A182W9E4 A0A1A9XUK9 A0A0T6BDY8 A0A336LY44 A0A1B0FIC5 A0A1B0AMG4 A0A0P4YVS4 A0A0P5G713
A0A0J7KTZ3 F4W6K3 E2AJX3 E2C022 A0A195BUV6 A0A158P2A8 A0A1I8QED3 A0A151X247 A0A0K8WB47 A0A034WIK9 A0A1A9Y1R7 A0A2J7QBX1 A0A2J7QBV5 A0A2J7QBV4 A0A182ILY7 A0A182P1S9 A0A1A9ZCE6 C3XZQ0 A0A182W9E4 A0A1A9XUK9 A0A0T6BDY8 A0A336LY44 A0A1B0FIC5 A0A1B0AMG4 A0A0P4YVS4 A0A0P5G713
PDB
4L6Y
E-value=6.85616e-39,
Score=406
Ontologies
PANTHER
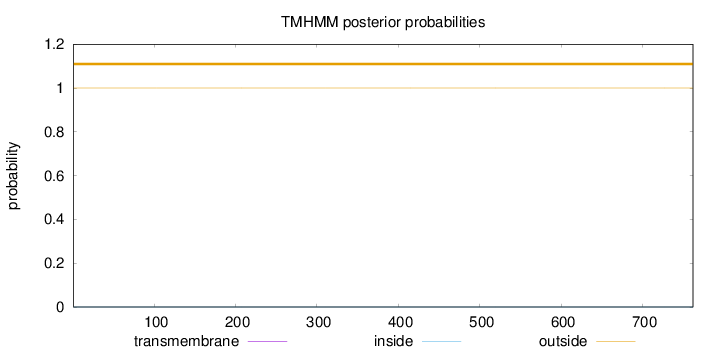
Topology
Length:
762
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00016
outside
1 - 762
Population Genetic Test Statistics
Pi
255.680671
Theta
189.276089
Tajima's D
1.179768
CLR
0.206603
CSRT
0.704664766761662
Interpretation
Uncertain
