Gene
KWMTBOMO10852
Annotation
PREDICTED:_protein_crooked_neck_[Bombyx_mori]
Full name
Protein crooked neck
Location in the cell
Cytoplasmic Reliability : 3.635
Sequence
CDS
ATGCCGAAAGTGGCAAAGGTGAAGAATAAAGCACCAGCAGAAATCCAAATAACAGCTGAACAATTACTTAGGGAAGCCAAAGAACGAGACCTTGAGATATTACCACCGCCACCTAAACAGAAAATATCTGATCCTGAGGAACTTCGAGACTACCAACATCGTAAACGTAAAGCATTTGAGGACAACATAAGAAAGAACAGACTTGTTATTGGGAACTGGCTTAAATATGCACAATGGGAAGAATCACAAAAGCAAATCCAAAGAGCTCGTTCAATATATGAGCGTGCTTTGGATGTGGATCACAGAAATGTTACCTTATGGCTGAAGTATACTGAAATGGAAATGAGGAACAGACAGGTAAACCATGCCCGTAATCTATGGGACAGAGCTGTTACAATTCTGCCAAGAGTATCTCAGTTTTGGTACAAATATACTTACATGGAGGAAATGTTGGAAAATGTAGCTGGAGCAAGACAGGTATTTGAACGTTGGATGGAATGGCATCCAGATGAGCAAGCTTGGCAAACATACATAAACTTTGAATTGAGGTACAAAGAACTTGAACGGGCTAGACAAATCTATGAACAGTTTGTTATGGTGCATCCAGATGTAAAGAATTGGATAAAATATGCAAAATTTGAAGAAAACCATGGTTTCATAAATGGCTCTAGGACAGTTTTTGAGAGAGCTGTTGAATTCTTTGGTGATGAAGAATTAGATGAAAGACTGTTTATAGCGTTTGCCAAATTTGAAGAAAATCAAAAAGAACATGACAGAGCAAGAGTGATTTACAAATATGCTCTGGATCATGTCCCAAAAGACAGAAATAAAGAACTTTATAAAGCTTATACTATTCATGAAAAGAAGTATGGTGATAGATCTGGCATTGAGGATGTAATAGTAAATAAGAGAAAGTATATGTATGAACAGGAAGTTATAGAGAATCCCACAAATTATGATGCATGGTTTGATTATATTAGATTAGTGGAGAATGAGGGTAATTTAGATGACATAAGAGACACATATGAGAGAGCAATTGCAAATATCCCTCCTTCAAAGGACAAACAATTTTGGAGGCGTTATATTTACTTGTGGATTAATTACGCTTTATATGAAGAGTTAGAAGCAGAAGATTTAGATCGAACAAGACAAGTTTACAGAACCTGTTTAGAATTAATTCCTCATAAGATATTTACTTTCTCAAAAATATGGCTCATGTATGCTCAGTTTGAAGTGAGATGTAAAGATTTAAAGCAAGCAAGAAAAACACTTGGTATGGCTCTGGGCATGTGCCCCAGAGATAAATTATACAGAGGTTATATAGACCTTGAGATACAGTTGAGAGAATTTGACAGATGTAGAATATTATACCAGAAATTCTTAGAGTATGGTCCAGAAAATTGTGTAACATGGATAAAGTTTGCAGAGTTAGAAACTCTGCTTGGAGATACAGATCGCGCAAGGGCTATTTATGAAATAGCTGTAGGTCAACAGAGATTAGATATGCCAGAATTGTTGTGGAAAAGCTATATCGATTTTGAGGTATCACAAGGAGAAACTGAAAAAGCAAGACAGCTTTATGAAAGACTTTTGGAACGAACAGTTCATGTCAAAGTGTGGCTTTCATATGCTAAATTTGAATTGAATGCTGAAAATGCTGATAATATTAATGTTGAATTGGCAAGACGGGTATATGAGCGTGCTAATGATAGTCTAAGAAGTGCAGGCGAAAAAGAATCGAGAGTTCTTCTCTTAGAAGCATGGAAGGAATTTGAAACAGAAATCGGAGATGAAGTGAAACTTGAAAAAGTCCTTGCCAAAATGCCTAGACGTGTCAAGAAGAGGCAGAAGATTATTAGTGAAGCTGGTGTTGAAGAAGGCTGGGAAGAAGTATTTGATTATATATTTCCAGAAGATGAAATGGTTAGACCTAATCTTAAATTGCTTGCTGCAGCTAAGAACTGGCGCAAACAACAAGAAATACCTCAAAGTACACAAAATGAAAAGGATGATAATGATGAGGAACCAGGAACCCCTGAAGGACATCCTACACAAGATAGTTCAAGTAATGATGGGTCAACAACGGCAGAATAA
Protein
MPKVAKVKNKAPAEIQITAEQLLREAKERDLEILPPPPKQKISDPEELRDYQHRKRKAFEDNIRKNRLVIGNWLKYAQWEESQKQIQRARSIYERALDVDHRNVTLWLKYTEMEMRNRQVNHARNLWDRAVTILPRVSQFWYKYTYMEEMLENVAGARQVFERWMEWHPDEQAWQTYINFELRYKELERARQIYEQFVMVHPDVKNWIKYAKFEENHGFINGSRTVFERAVEFFGDEELDERLFIAFAKFEENQKEHDRARVIYKYALDHVPKDRNKELYKAYTIHEKKYGDRSGIEDVIVNKRKYMYEQEVIENPTNYDAWFDYIRLVENEGNLDDIRDTYERAIANIPPSKDKQFWRRYIYLWINYALYEELEAEDLDRTRQVYRTCLELIPHKIFTFSKIWLMYAQFEVRCKDLKQARKTLGMALGMCPRDKLYRGYIDLEIQLREFDRCRILYQKFLEYGPENCVTWIKFAELETLLGDTDRARAIYEIAVGQQRLDMPELLWKSYIDFEVSQGETEKARQLYERLLERTVHVKVWLSYAKFELNAENADNINVELARRVYERANDSLRSAGEKESRVLLLEAWKEFETEIGDEVKLEKVLAKMPRRVKKRQKIISEAGVEEGWEEVFDYIFPEDEMVRPNLKLLAAAKNWRKQQEIPQSTQNEKDDNDEEPGTPEGHPTQDSSSNDGSTTAE
Summary
Description
May be involved in pre-mRNA splicing process. Involved in neurogenesis.
Subunit
Colocalizes with a complex containing snRNP proteins.
Similarity
Belongs to the crooked-neck family.
Keywords
Complete proteome
Developmental protein
Differentiation
mRNA processing
mRNA splicing
Neurogenesis
Nucleus
Reference proteome
Repeat
Feature
chain Protein crooked neck
Uniprot
A0A2A4JE21
A0A088ML72
A0A291P118
A0A291P120
A0A1V0M8A8
A0A0P0EUP4
+ More
A0A2H1VMH2 A0A194PH50 A0A194QNM9 Q16W00 A0A182H3J2 A0A1I8N6A3 B0WMF3 A0A1Q3F2Y2 A0A0L0CCU8 A0A182UQY5 A0A182MWE0 A0A182VQW6 A0A2M4AQX0 A0A182YJV6 A0A182ULD6 A0A084VRV0 A0A182KM51 Q7PYD1 A0A182XBN0 A0A336MQE5 A0A182I0G6 A0A182JUL4 W8BPA9 A0A182R1H5 A0A182J8E7 A0A182N4W3 W5J4T2 A0A182QSC2 A0A182PEE6 A0A182FQL6 A0A182T8D3 A0A0K8V4Q4 A0A0A1X599 A0A034WVH3 A0A1I8NZM9 A0A0T6AU33 A0A0N0BKQ9 A0A088AAB8 A0A1A9WX46 A0A2A3E4B3 A0A1B0FPF4 A0A1B0BFU1 A0A1B0AC07 A0A1B0BVZ5 A0A1A9ULF4 B4Q182 A0A1B6FPF1 D6X558 A0A1A9XSZ8 B3P8Z5 A0A1W4V213 A0A1Y1KBA4 A0A1A9Y1R5 A0A3B0K5J9 B4I9T0 P17886 B3MYT2 B4L5U5 B5DKF4 A0A0M3QZJ6 A0A1B6D9W9 E9IV64 E0VFZ4 B4M7D3 B4JJ81 U4U2G7 T1HM51 A0A026VVA0 A0A0V0G855 A0A3L8D4I3 A0A232EP28 B4N1M0 K7IZB8 A0A1J1IHK3 A0A2J7Q660 E2AIS0 A0A0L7QNZ2 A0A195CPN5 F4WVK3 E2B3I6 A0A0J7L5Q8 A0A195BV78 A0A158NEF5 A0A195EP64 A0A195FKB5 B4H364 A0A1A9VSD0 A0A0C9RJP8 A0A0A9XKU8 A0A2J7PL27 L7MIM2 A0A023GGB1 A0A224YVE5 A0A151XJ98
A0A2H1VMH2 A0A194PH50 A0A194QNM9 Q16W00 A0A182H3J2 A0A1I8N6A3 B0WMF3 A0A1Q3F2Y2 A0A0L0CCU8 A0A182UQY5 A0A182MWE0 A0A182VQW6 A0A2M4AQX0 A0A182YJV6 A0A182ULD6 A0A084VRV0 A0A182KM51 Q7PYD1 A0A182XBN0 A0A336MQE5 A0A182I0G6 A0A182JUL4 W8BPA9 A0A182R1H5 A0A182J8E7 A0A182N4W3 W5J4T2 A0A182QSC2 A0A182PEE6 A0A182FQL6 A0A182T8D3 A0A0K8V4Q4 A0A0A1X599 A0A034WVH3 A0A1I8NZM9 A0A0T6AU33 A0A0N0BKQ9 A0A088AAB8 A0A1A9WX46 A0A2A3E4B3 A0A1B0FPF4 A0A1B0BFU1 A0A1B0AC07 A0A1B0BVZ5 A0A1A9ULF4 B4Q182 A0A1B6FPF1 D6X558 A0A1A9XSZ8 B3P8Z5 A0A1W4V213 A0A1Y1KBA4 A0A1A9Y1R5 A0A3B0K5J9 B4I9T0 P17886 B3MYT2 B4L5U5 B5DKF4 A0A0M3QZJ6 A0A1B6D9W9 E9IV64 E0VFZ4 B4M7D3 B4JJ81 U4U2G7 T1HM51 A0A026VVA0 A0A0V0G855 A0A3L8D4I3 A0A232EP28 B4N1M0 K7IZB8 A0A1J1IHK3 A0A2J7Q660 E2AIS0 A0A0L7QNZ2 A0A195CPN5 F4WVK3 E2B3I6 A0A0J7L5Q8 A0A195BV78 A0A158NEF5 A0A195EP64 A0A195FKB5 B4H364 A0A1A9VSD0 A0A0C9RJP8 A0A0A9XKU8 A0A2J7PL27 L7MIM2 A0A023GGB1 A0A224YVE5 A0A151XJ98
Pubmed
26385554
28756777
28986331
28349297
26445454
26354079
+ More
17510324 26483478 25315136 26108605 25244985 24438588 20966253 12364791 14747013 17210077 24495485 20920257 23761445 25830018 25348373 17994087 17550304 18362917 19820115 28004739 2044955 10731132 12537572 10731137 12537569 12163015 15632085 21282665 20566863 23537049 24508170 30249741 28648823 20075255 20798317 21719571 21347285 25401762 26823975 25576852 28797301
17510324 26483478 25315136 26108605 25244985 24438588 20966253 12364791 14747013 17210077 24495485 20920257 23761445 25830018 25348373 17994087 17550304 18362917 19820115 28004739 2044955 10731132 12537572 10731137 12537569 12163015 15632085 21282665 20566863 23537049 24508170 30249741 28648823 20075255 20798317 21719571 21347285 25401762 26823975 25576852 28797301
EMBL
NWSH01001729
PCG70325.1
KJ579209
AIN34685.1
MF706170
KZ150058
+ More
ATJ44597.1 PZC74278.1 MF687642 ATJ44568.1 KU755511 ARD71221.1 KT261713 ALJ30253.1 ODYU01003204 SOQ41642.1 KQ459604 KPI92627.1 KQ461196 KPJ06580.1 CH477578 EAT38766.1 JXUM01003845 KQ560164 KXJ84073.1 DS231997 EDS31010.1 GFDL01013135 JAV21910.1 JRES01000586 KNC30037.1 AXCM01005019 GGFK01009865 MBW43186.1 ATLV01015780 KE525036 KFB40694.1 AAAB01008987 EAA01065.4 UFQT01000960 SSX28198.1 APCN01002077 GAMC01003585 JAC02971.1 ADMH02002134 ETN58403.1 AXCN02000188 GDHF01018392 JAI33922.1 GBXI01008216 JAD06076.1 GAKP01001184 JAC57768.1 LJIG01022870 KRT78302.1 KQ435695 KOX80872.1 KZ288392 PBC26334.1 CCAG010016287 JXJN01013668 JXJN01021549 CM000162 EDX01389.1 GECZ01029103 GECZ01017876 GECZ01015322 GECZ01005754 JAS40666.1 JAS51893.1 JAS54447.1 JAS64015.1 KQ971382 EEZ97205.1 CH954183 EDV45600.1 GEZM01087319 JAV58773.1 OUUW01000003 SPP78728.1 CH480825 EDW43961.1 X58374 AE014298 AL009195 AY051666 CH902632 EDV32776.1 CH933811 EDW06554.1 CH379063 EDY72003.1 CP012528 ALC49533.1 GEDC01014805 JAS22493.1 GL766135 EFZ15538.1 DS235129 EEB12300.1 CH940653 EDW62700.1 CH916370 EDV99633.1 KB631815 ERL86498.1 ACPB03008137 KK107796 EZA47662.1 GECL01001918 JAP04206.1 QOIP01000013 RLU15146.1 NNAY01003030 OXU20091.1 CH963925 EDW78259.1 CVRI01000052 CRK99733.1 NEVH01017540 PNF24062.1 GL439869 EFN66682.1 KQ414843 KOC60338.1 KQ977444 KYN02671.1 GL888387 EGI61824.1 GL445346 EFN89738.1 LBMM01000651 KMQ97888.1 KQ976401 KYM92487.1 ADTU01013276 KQ978625 KYN29674.1 KQ981490 KYN41110.1 CH479205 EDW30757.1 GBYB01013497 GBYB01013498 JAG83264.1 JAG83265.1 GBHO01022252 GDHC01014210 JAG21352.1 JAQ04419.1 NEVH01024529 PNF17019.1 GACK01001194 JAA63840.1 GBBM01003618 JAC31800.1 GFPF01006656 MAA17802.1 KQ982080 KYQ60407.1
ATJ44597.1 PZC74278.1 MF687642 ATJ44568.1 KU755511 ARD71221.1 KT261713 ALJ30253.1 ODYU01003204 SOQ41642.1 KQ459604 KPI92627.1 KQ461196 KPJ06580.1 CH477578 EAT38766.1 JXUM01003845 KQ560164 KXJ84073.1 DS231997 EDS31010.1 GFDL01013135 JAV21910.1 JRES01000586 KNC30037.1 AXCM01005019 GGFK01009865 MBW43186.1 ATLV01015780 KE525036 KFB40694.1 AAAB01008987 EAA01065.4 UFQT01000960 SSX28198.1 APCN01002077 GAMC01003585 JAC02971.1 ADMH02002134 ETN58403.1 AXCN02000188 GDHF01018392 JAI33922.1 GBXI01008216 JAD06076.1 GAKP01001184 JAC57768.1 LJIG01022870 KRT78302.1 KQ435695 KOX80872.1 KZ288392 PBC26334.1 CCAG010016287 JXJN01013668 JXJN01021549 CM000162 EDX01389.1 GECZ01029103 GECZ01017876 GECZ01015322 GECZ01005754 JAS40666.1 JAS51893.1 JAS54447.1 JAS64015.1 KQ971382 EEZ97205.1 CH954183 EDV45600.1 GEZM01087319 JAV58773.1 OUUW01000003 SPP78728.1 CH480825 EDW43961.1 X58374 AE014298 AL009195 AY051666 CH902632 EDV32776.1 CH933811 EDW06554.1 CH379063 EDY72003.1 CP012528 ALC49533.1 GEDC01014805 JAS22493.1 GL766135 EFZ15538.1 DS235129 EEB12300.1 CH940653 EDW62700.1 CH916370 EDV99633.1 KB631815 ERL86498.1 ACPB03008137 KK107796 EZA47662.1 GECL01001918 JAP04206.1 QOIP01000013 RLU15146.1 NNAY01003030 OXU20091.1 CH963925 EDW78259.1 CVRI01000052 CRK99733.1 NEVH01017540 PNF24062.1 GL439869 EFN66682.1 KQ414843 KOC60338.1 KQ977444 KYN02671.1 GL888387 EGI61824.1 GL445346 EFN89738.1 LBMM01000651 KMQ97888.1 KQ976401 KYM92487.1 ADTU01013276 KQ978625 KYN29674.1 KQ981490 KYN41110.1 CH479205 EDW30757.1 GBYB01013497 GBYB01013498 JAG83264.1 JAG83265.1 GBHO01022252 GDHC01014210 JAG21352.1 JAQ04419.1 NEVH01024529 PNF17019.1 GACK01001194 JAA63840.1 GBBM01003618 JAC31800.1 GFPF01006656 MAA17802.1 KQ982080 KYQ60407.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000008820
UP000069940
UP000249989
+ More
UP000095301 UP000002320 UP000037069 UP000075903 UP000075883 UP000075920 UP000076408 UP000075902 UP000030765 UP000075882 UP000007062 UP000076407 UP000075840 UP000075881 UP000075900 UP000075880 UP000075884 UP000000673 UP000075886 UP000075885 UP000069272 UP000075901 UP000095300 UP000053105 UP000005203 UP000091820 UP000242457 UP000092444 UP000092460 UP000092445 UP000078200 UP000002282 UP000007266 UP000092443 UP000008711 UP000192221 UP000268350 UP000001292 UP000000803 UP000007801 UP000009192 UP000001819 UP000092553 UP000009046 UP000008792 UP000001070 UP000030742 UP000015103 UP000053097 UP000279307 UP000215335 UP000007798 UP000002358 UP000183832 UP000235965 UP000000311 UP000053825 UP000078542 UP000007755 UP000008237 UP000036403 UP000078540 UP000005205 UP000078492 UP000078541 UP000008744 UP000075809
UP000095301 UP000002320 UP000037069 UP000075903 UP000075883 UP000075920 UP000076408 UP000075902 UP000030765 UP000075882 UP000007062 UP000076407 UP000075840 UP000075881 UP000075900 UP000075880 UP000075884 UP000000673 UP000075886 UP000075885 UP000069272 UP000075901 UP000095300 UP000053105 UP000005203 UP000091820 UP000242457 UP000092444 UP000092460 UP000092445 UP000078200 UP000002282 UP000007266 UP000092443 UP000008711 UP000192221 UP000268350 UP000001292 UP000000803 UP000007801 UP000009192 UP000001819 UP000092553 UP000009046 UP000008792 UP000001070 UP000030742 UP000015103 UP000053097 UP000279307 UP000215335 UP000007798 UP000002358 UP000183832 UP000235965 UP000000311 UP000053825 UP000078542 UP000007755 UP000008237 UP000036403 UP000078540 UP000005205 UP000078492 UP000078541 UP000008744 UP000075809
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2A4JE21
A0A088ML72
A0A291P118
A0A291P120
A0A1V0M8A8
A0A0P0EUP4
+ More
A0A2H1VMH2 A0A194PH50 A0A194QNM9 Q16W00 A0A182H3J2 A0A1I8N6A3 B0WMF3 A0A1Q3F2Y2 A0A0L0CCU8 A0A182UQY5 A0A182MWE0 A0A182VQW6 A0A2M4AQX0 A0A182YJV6 A0A182ULD6 A0A084VRV0 A0A182KM51 Q7PYD1 A0A182XBN0 A0A336MQE5 A0A182I0G6 A0A182JUL4 W8BPA9 A0A182R1H5 A0A182J8E7 A0A182N4W3 W5J4T2 A0A182QSC2 A0A182PEE6 A0A182FQL6 A0A182T8D3 A0A0K8V4Q4 A0A0A1X599 A0A034WVH3 A0A1I8NZM9 A0A0T6AU33 A0A0N0BKQ9 A0A088AAB8 A0A1A9WX46 A0A2A3E4B3 A0A1B0FPF4 A0A1B0BFU1 A0A1B0AC07 A0A1B0BVZ5 A0A1A9ULF4 B4Q182 A0A1B6FPF1 D6X558 A0A1A9XSZ8 B3P8Z5 A0A1W4V213 A0A1Y1KBA4 A0A1A9Y1R5 A0A3B0K5J9 B4I9T0 P17886 B3MYT2 B4L5U5 B5DKF4 A0A0M3QZJ6 A0A1B6D9W9 E9IV64 E0VFZ4 B4M7D3 B4JJ81 U4U2G7 T1HM51 A0A026VVA0 A0A0V0G855 A0A3L8D4I3 A0A232EP28 B4N1M0 K7IZB8 A0A1J1IHK3 A0A2J7Q660 E2AIS0 A0A0L7QNZ2 A0A195CPN5 F4WVK3 E2B3I6 A0A0J7L5Q8 A0A195BV78 A0A158NEF5 A0A195EP64 A0A195FKB5 B4H364 A0A1A9VSD0 A0A0C9RJP8 A0A0A9XKU8 A0A2J7PL27 L7MIM2 A0A023GGB1 A0A224YVE5 A0A151XJ98
A0A2H1VMH2 A0A194PH50 A0A194QNM9 Q16W00 A0A182H3J2 A0A1I8N6A3 B0WMF3 A0A1Q3F2Y2 A0A0L0CCU8 A0A182UQY5 A0A182MWE0 A0A182VQW6 A0A2M4AQX0 A0A182YJV6 A0A182ULD6 A0A084VRV0 A0A182KM51 Q7PYD1 A0A182XBN0 A0A336MQE5 A0A182I0G6 A0A182JUL4 W8BPA9 A0A182R1H5 A0A182J8E7 A0A182N4W3 W5J4T2 A0A182QSC2 A0A182PEE6 A0A182FQL6 A0A182T8D3 A0A0K8V4Q4 A0A0A1X599 A0A034WVH3 A0A1I8NZM9 A0A0T6AU33 A0A0N0BKQ9 A0A088AAB8 A0A1A9WX46 A0A2A3E4B3 A0A1B0FPF4 A0A1B0BFU1 A0A1B0AC07 A0A1B0BVZ5 A0A1A9ULF4 B4Q182 A0A1B6FPF1 D6X558 A0A1A9XSZ8 B3P8Z5 A0A1W4V213 A0A1Y1KBA4 A0A1A9Y1R5 A0A3B0K5J9 B4I9T0 P17886 B3MYT2 B4L5U5 B5DKF4 A0A0M3QZJ6 A0A1B6D9W9 E9IV64 E0VFZ4 B4M7D3 B4JJ81 U4U2G7 T1HM51 A0A026VVA0 A0A0V0G855 A0A3L8D4I3 A0A232EP28 B4N1M0 K7IZB8 A0A1J1IHK3 A0A2J7Q660 E2AIS0 A0A0L7QNZ2 A0A195CPN5 F4WVK3 E2B3I6 A0A0J7L5Q8 A0A195BV78 A0A158NEF5 A0A195EP64 A0A195FKB5 B4H364 A0A1A9VSD0 A0A0C9RJP8 A0A0A9XKU8 A0A2J7PL27 L7MIM2 A0A023GGB1 A0A224YVE5 A0A151XJ98
PDB
6QDV
E-value=0,
Score=2223
Ontologies
GO
Topology
Subcellular location
Nucleus speckle
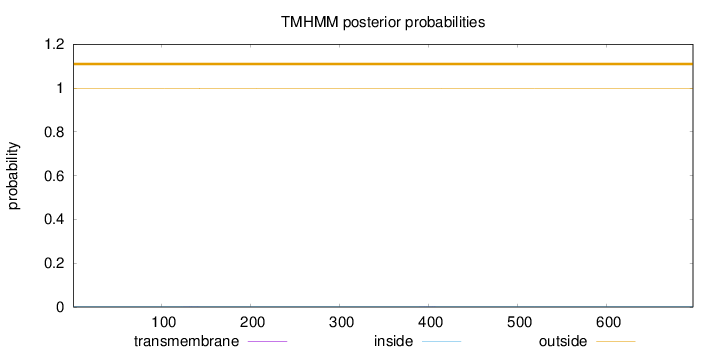
Length:
697
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00924999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00150
outside
1 - 697
Population Genetic Test Statistics
Pi
35.588419
Theta
155.893835
Tajima's D
-1.684172
CLR
2512.530901
CSRT
0.039698015099245
Interpretation
Possibly Positive selection
