Gene
KWMTBOMO10849
Pre Gene Modal
BGIBMGA008457
Annotation
PREDICTED:_diphthine--ammonia_ligase_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 3.232
Sequence
CDS
ATGAGAACTGTAGCGTTAATAAGTGGTGGCAAAGACAGCTGCTACAACATGTTGCAATGTGTAGGGGCCGGGCACACTATCGTTGCACTGGCGAATCTCCAACCTCTTCATAAAGATGAATTGGACAGTTACATGTATCAATCAGTAGGTCATCATGGCATAGAACTTTATGCTGAAGCAATGGGATTACCTCTTTATAGAGAAATAATAACAGGTACAGCTGTAGATCAAGGAAAAAATTACAAACCCACTGATAATGATGAAGTTGAGGACCTATACAGGTTGCTTTCCAAAGTCAAGGAAGAAATAAATATAGAAGCAGTTGCTGTTGGTGCAATACTATCTGACTACCAAAGAATCAGAGTTGAGAATGTATGTCAACGCTTGGGTCTAGTTTCACTCGCTTATTTATGGAGGCGGAACCAAAAAGAGTTGTTACAGGAAATGATATACTGTGGTATTGATGCGATCATTATAAAGGTGGCAGCTATGGGTCTCGATCCGAGGATACACTTAGGTATGACGATACGCGACATCCAGCCACATTTGCTAGTCATGAAAGAGAAGTACGGGCTCAACATGTGCGGGGAGGGGGGAGAGTACGAAACCTTCACACTCGACTGTCCCCTGTTCAAGAAAAAACTGATCATCGATGAAAAAGAAATGGTTATGCATTCAGACGATCCTGTAGCTCCTGTCGGGTATTTGAACTTGAAATTGCACCTCGAGCCTAAAGAGAACTGCGACGAACCTCTCCAGCTACCTCCAATCATTAAAAACTCGTTAGACTATTTAAACGACTTGAGCGATTCGGTCTTTAGCGACATAAGCGATATCGAATTGAGTGAGACAGAAATAGAGTCTATAGAGAAGTTAGAGAAAGAGGAAAAGATGAAGAACAGTGCCGAACTCAATCCGTTGCTCACTTATTCGCCGGACAAAGAGAACAATTTAGATATTATACACAAGTTAAATGATAGTGCGGAAGTATTAGACAACGGATATGTATACGAACAGACGAACGATGATAGGTCGTCGCACACGATAACGGATCATCGATCGATATACGGGAACAAACATGGATGGTATTTTATTGGAGGGATTGTCGGCGAAGGGATCGACACGAGGGTGGCAACTGAAGATGCGATGAATAAATTAATTAGTTTGTTAGATTCTGAAAGCATGCATCTAACCGATGTGTGCTCCGTCAACATATACATGAGGAATATGGAAGAGTATGCTTCGTTGAATGACGTTTATGTCAAGACGTTTTGCTACCCGAACCCCCCGACGCGTGTCTGCGTACAGTGCCCTCTGCCGAACAACGTAGGACTTATTTTGGATGGGGTGGCGTTCAAATCAAGTAACAACAATACCGAGACTGACGGAGGTAACGTGAAGGTGCGAGAGACTATGCATGTTCAAGGCATTTCGCATTGGGCCCCAGCCAACATCGGACCTTATAGTCAAGCGATTAAGGTGGGTGAAGTAATCGGAACGTGCGGTCAGATAGCGTTAATACCGGGCGTCATGACGCTGGCGGGCGGCGGCGCGCGCAGACAGAGCGCGCTCGCGCTGCGACACCTCACGCGCGTCATACGGGCGGCGCATCCACGGGCGCACATACGTAGCGTCGTACAGGTAATATAG
Protein
MRTVALISGGKDSCYNMLQCVGAGHTIVALANLQPLHKDELDSYMYQSVGHHGIELYAEAMGLPLYREIITGTAVDQGKNYKPTDNDEVEDLYRLLSKVKEEINIEAVAVGAILSDYQRIRVENVCQRLGLVSLAYLWRRNQKELLQEMIYCGIDAIIIKVAAMGLDPRIHLGMTIRDIQPHLLVMKEKYGLNMCGEGGEYETFTLDCPLFKKKLIIDEKEMVMHSDDPVAPVGYLNLKLHLEPKENCDEPLQLPPIIKNSLDYLNDLSDSVFSDISDIELSETEIESIEKLEKEEKMKNSAELNPLLTYSPDKENNLDIIHKLNDSAEVLDNGYVYEQTNDDRSSHTITDHRSIYGNKHGWYFIGGIVGEGIDTRVATEDAMNKLISLLDSESMHLTDVCSVNIYMRNMEEYASLNDVYVKTFCYPNPPTRVCVQCPLPNNVGLILDGVAFKSSNNNTETDGGNVKVRETMHVQGISHWAPANIGPYSQAIKVGEVIGTCGQIALIPGVMTLAGGGARRQSALALRHLTRVIRAAHPRAHIRSVVQVI
Summary
Uniprot
H9JG10
A0A2H1WN34
A0A2A4KA83
A0A2W1BKW1
A0A2A4K8M5
A0A212EW48
+ More
A0A194PND4 A0A194QNN4 A0A067RNS0 A0A2J7PL03 A0A1J1IFH5 A0A1Q3EU54 A0A084VS41 A0A182J5Q0 A0A154P2F8 K7J9Q7 A0A2A3EE91 A0A088AM37 E2BW99 A0A0K8TRG8 A0A1B6EUR4 A0A2S2PXD6 D2A2B0 A0A232EVN6 A0A1B6HEN6 E2AII7 A0A0T6B8E1 A0A1S3J8G2 A0A087YAW6 A0A1A8UT71 W5N314 A0A1A8ITU1 A0A096LQQ1 A0A1A8RZV3 W5N319 A0A1A8LE73 A0A3B4BG83 K7FL01 V9KG05 A0A397G8J8
A0A194PND4 A0A194QNN4 A0A067RNS0 A0A2J7PL03 A0A1J1IFH5 A0A1Q3EU54 A0A084VS41 A0A182J5Q0 A0A154P2F8 K7J9Q7 A0A2A3EE91 A0A088AM37 E2BW99 A0A0K8TRG8 A0A1B6EUR4 A0A2S2PXD6 D2A2B0 A0A232EVN6 A0A1B6HEN6 E2AII7 A0A0T6B8E1 A0A1S3J8G2 A0A087YAW6 A0A1A8UT71 W5N314 A0A1A8ITU1 A0A096LQQ1 A0A1A8RZV3 W5N319 A0A1A8LE73 A0A3B4BG83 K7FL01 V9KG05 A0A397G8J8
Pubmed
EMBL
BABH01001793
BABH01001794
ODYU01009812
SOQ54481.1
NWSH01000028
PCG80562.1
+ More
KZ150058 PZC74274.1 PCG80561.1 AGBW02012095 OWR45687.1 KQ459604 KPI92630.1 KQ461196 KPJ06585.1 KK852559 KDR21389.1 NEVH01024529 PNF17013.1 CVRI01000048 CRK99013.1 GFDL01016200 JAV18845.1 ATLV01015797 ATLV01015798 KE525036 KFB40785.1 KQ434806 KZC06125.1 AAZX01005119 KZ288282 PBC29539.1 GL451103 EFN80003.1 GDAI01001088 JAI16515.1 GECZ01028306 JAS41463.1 GGMS01000821 MBY70024.1 KQ971338 EFA02050.1 NNAY01001974 OXU22410.1 GECU01034601 GECU01016534 GECU01010007 JAS73105.1 JAS91172.1 JAS97699.1 GL439791 EFN66777.1 LJIG01009227 KRT83502.1 AYCK01001363 AYCK01001364 HAEJ01011112 SBS51569.1 AHAT01019067 AHAT01019068 HAED01014316 SBR00761.1 HAEH01020460 SBS10823.1 HAEF01004797 SBR42179.1 AGCU01150533 AGCU01150534 AGCU01150535 AGCU01150536 AGCU01150537 AGCU01150538 AGCU01150539 AGCU01150540 AGCU01150541 AGCU01150542 JW864292 AFO96809.1 PQFF01000492 RHZ47352.1
KZ150058 PZC74274.1 PCG80561.1 AGBW02012095 OWR45687.1 KQ459604 KPI92630.1 KQ461196 KPJ06585.1 KK852559 KDR21389.1 NEVH01024529 PNF17013.1 CVRI01000048 CRK99013.1 GFDL01016200 JAV18845.1 ATLV01015797 ATLV01015798 KE525036 KFB40785.1 KQ434806 KZC06125.1 AAZX01005119 KZ288282 PBC29539.1 GL451103 EFN80003.1 GDAI01001088 JAI16515.1 GECZ01028306 JAS41463.1 GGMS01000821 MBY70024.1 KQ971338 EFA02050.1 NNAY01001974 OXU22410.1 GECU01034601 GECU01016534 GECU01010007 JAS73105.1 JAS91172.1 JAS97699.1 GL439791 EFN66777.1 LJIG01009227 KRT83502.1 AYCK01001363 AYCK01001364 HAEJ01011112 SBS51569.1 AHAT01019067 AHAT01019068 HAED01014316 SBR00761.1 HAEH01020460 SBS10823.1 HAEF01004797 SBR42179.1 AGCU01150533 AGCU01150534 AGCU01150535 AGCU01150536 AGCU01150537 AGCU01150538 AGCU01150539 AGCU01150540 AGCU01150541 AGCU01150542 JW864292 AFO96809.1 PQFF01000492 RHZ47352.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF55298
SSF55298
Gene 3D
ProteinModelPortal
H9JG10
A0A2H1WN34
A0A2A4KA83
A0A2W1BKW1
A0A2A4K8M5
A0A212EW48
+ More
A0A194PND4 A0A194QNN4 A0A067RNS0 A0A2J7PL03 A0A1J1IFH5 A0A1Q3EU54 A0A084VS41 A0A182J5Q0 A0A154P2F8 K7J9Q7 A0A2A3EE91 A0A088AM37 E2BW99 A0A0K8TRG8 A0A1B6EUR4 A0A2S2PXD6 D2A2B0 A0A232EVN6 A0A1B6HEN6 E2AII7 A0A0T6B8E1 A0A1S3J8G2 A0A087YAW6 A0A1A8UT71 W5N314 A0A1A8ITU1 A0A096LQQ1 A0A1A8RZV3 W5N319 A0A1A8LE73 A0A3B4BG83 K7FL01 V9KG05 A0A397G8J8
A0A194PND4 A0A194QNN4 A0A067RNS0 A0A2J7PL03 A0A1J1IFH5 A0A1Q3EU54 A0A084VS41 A0A182J5Q0 A0A154P2F8 K7J9Q7 A0A2A3EE91 A0A088AM37 E2BW99 A0A0K8TRG8 A0A1B6EUR4 A0A2S2PXD6 D2A2B0 A0A232EVN6 A0A1B6HEN6 E2AII7 A0A0T6B8E1 A0A1S3J8G2 A0A087YAW6 A0A1A8UT71 W5N314 A0A1A8ITU1 A0A096LQQ1 A0A1A8RZV3 W5N319 A0A1A8LE73 A0A3B4BG83 K7FL01 V9KG05 A0A397G8J8
PDB
3RK1
E-value=1.10151e-29,
Score=325
Ontologies
PANTHER
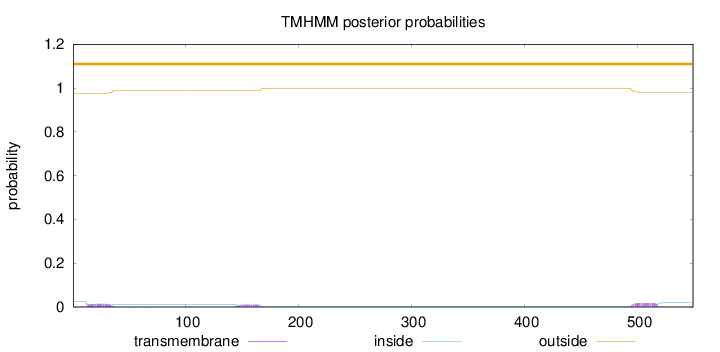
Topology
Length:
549
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.92592
Exp number, first 60 AAs:
0.29464
Total prob of N-in:
0.02411
outside
1 - 549
Population Genetic Test Statistics
Pi
255.002548
Theta
174.311801
Tajima's D
0.862318
CLR
0.045017
CSRT
0.619269036548173
Interpretation
Uncertain
