Gene
KWMTBOMO10848
Pre Gene Modal
BGIBMGA008364
Annotation
PREDICTED:_REST_corepressor_isoform_X3_[Papilio_xuthus]
Full name
REST corepressor
Alternative Name
CoREST
Transcription factor
Location in the cell
Nuclear Reliability : 3.523
Sequence
CDS
ATGGTTCTAGCAGAACGAAGTAATGATGTTCGAAACGGGAAGCGGTCAAGGGGGCCAAGCCCCAATGGTCATGGAAGTCCAGATTCAAGTTCGGATGAAGAAAATGTAGTACCGTTTGCAGCCGAAAAGATTCGGGTTGGTCGGGATTACCAAGCAGTCTGTCCAGAATTGGAACCAGTTGAAGCAAGAAGACCAGACTTAATTTCTGATAGGGCTCTGCTTGTCTGGTCGCCAACTACTGAGATAGCTGATATCAAATTGGATGAATATATTACAACAGCTAAAGAAAAATATGGTTATAATGGTGAACAAGCTCTTGGTATGCTCTTCTGGCACAAGCATGATCTTAATCGTGCATCATTGGATCTTGCAAACTTCACACCATTTCCTGATGAGTGGACAGTGGAGGACAAGGTGTTATTTGAACAAGCTTTTCAGTTTCATGGTAAAAGCTTTCATCGAATACGTCAAATGCTTCCAGATAAATCTATTGCATCTTTAGTCAAATACTATTATTCATGGAAGAAAACAAGAGCACGTACTTCCTTAATGGATGTTGTTGGTGAAGGGCGAAATGCTGCAAGCTCAGGGTCTGGAAAAAAAGAGTCTGGGACTACATCTGAACCTGGCGGCTCAGACAAAGAATCTGACAATGATGAAAAGGTACAACAAAACAAGCAGCAATTGAGCGCCATGAAACGCAAAGTTGGCGATACTGGAGTGGAGGATTACCGTCCTGGCGAGCCTCCTGCCAAGATAAATTCACGTTGGACAAATGACGAATTACTCATGGCCGTTACTGCAGTAAGAAAGTACGGTAAGGAACTTATCTTGGAGAACTCCCTTTGCTGTGGCGGTAACAAAATAAATTTTGTACGTGACAAATAA
Protein
MVLAERSNDVRNGKRSRGPSPNGHGSPDSSSDEENVVPFAAEKIRVGRDYQAVCPELEPVEARRPDLISDRALLVWSPTTEIADIKLDEYITTAKEKYGYNGEQALGMLFWHKHDLNRASLDLANFTPFPDEWTVEDKVLFEQAFQFHGKSFHRIRQMLPDKSIASLVKYYYSWKKTRARTSLMDVVGEGRNAASSGSGKKESGTTSEPGGSDKESDNDEKVQQNKQQLSAMKRKVGDTGVEDYRPGEPPAKINSRWTNDELLMAVTAVRKYGKELILENSLCCGGNKINFVRDK
Summary
Description
Essential component of a corepressor complex that represses transcription of neuron-specific genes in non-neuronal cells. The BHC complex is recruited by Ttk88 and probably acts by deacetylating and demethylating specific sites on histones, thereby acting as a chromatin modifier. May serve as a molecular beacon for the recruitment of molecular machinery that imposes silencing across a chromosomal interval.
Subunit
Component of a complex that contains at least Rpd3, CoRest and Su(var)3-3/Hdm. Interacts with neuronal repressor ttk/Ttk88.
Similarity
Belongs to the CoREST family.
Keywords
Alternative splicing
Chromatin regulator
Coiled coil
Complete proteome
Nucleus
Phosphoprotein
Reference proteome
Repeat
Repressor
Transcription
Transcription regulation
Feature
chain REST corepressor
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
A0A2A4K9A8
A0A2W1BR93
H9JFR7
A0A212EW08
A0A2A4K8H2
A0A2A4K8N8
+ More
A0A2H1WN35 A0A2A4K9J2 A0A194PGW2 D2A064 A0A1I8M1Q7 T1PBR0 A0A1Y1K5V9 A0A194QSS1 W8BQ39 A0A034W9G7 A0A0A1WL46 A0A0K8TKM1 A0A1I8NWR0 U5EQ84 A0A1L8DGE5 A0A0L7KRJ7 B3NVP2 B4PZH4 Q59E36 A0A1W4WQJ4 A0A1W4WPI9 A0A0N8P243 A0A1W4VUS2 Q17M84 B4MA37 A0A0Q9WP20 Q59E36-4 A0A3B0KAQ5 A0A1Y1KAM5 A0A139WJB2 W8BIL6 A0A0L7RI93 A0A1B0DG41 A0A182TW84 A0A182N535 Q7QC78 E2BWA1 W8CAT6 A0A1B6E6Q8 A0A182K191 A0A2J7PL06 A0A023EVK7 A0A195DWG3 A0A182GVN2 A0A1I8M1Q3 A0A182IM06 F4X132 A0A1B6E7D2 A0A182QVL8 A0A182VB33 A0A067RLA0 A0A0L0BRK8 A0A1B6CTF4 A0A182HSW8 A0A182RGY2 A0A182XHY7 A0A182VWQ3 A0A084VAN0 A0A1B6BZ51 A0A2M4AW31 A0A0A1XI16 A0A2M4ASF9 A0A0K8WJB9 A0A2M4AUG5 M9PJR2 A0A034W6U3 W8BW02 Q59E36-3 A0A0R1EAT3 A0A182MCU5 A0A1A9VWZ2 A0A182YHK4 A0A1B6K8A3 A0A1L8DG32 A0A0Q5TJS9 A0A0P9AX75 A0A1B0ASU1 A0A1B0FLM9 A0A034W8H3 A0A1A9XYG8
A0A2H1WN35 A0A2A4K9J2 A0A194PGW2 D2A064 A0A1I8M1Q7 T1PBR0 A0A1Y1K5V9 A0A194QSS1 W8BQ39 A0A034W9G7 A0A0A1WL46 A0A0K8TKM1 A0A1I8NWR0 U5EQ84 A0A1L8DGE5 A0A0L7KRJ7 B3NVP2 B4PZH4 Q59E36 A0A1W4WQJ4 A0A1W4WPI9 A0A0N8P243 A0A1W4VUS2 Q17M84 B4MA37 A0A0Q9WP20 Q59E36-4 A0A3B0KAQ5 A0A1Y1KAM5 A0A139WJB2 W8BIL6 A0A0L7RI93 A0A1B0DG41 A0A182TW84 A0A182N535 Q7QC78 E2BWA1 W8CAT6 A0A1B6E6Q8 A0A182K191 A0A2J7PL06 A0A023EVK7 A0A195DWG3 A0A182GVN2 A0A1I8M1Q3 A0A182IM06 F4X132 A0A1B6E7D2 A0A182QVL8 A0A182VB33 A0A067RLA0 A0A0L0BRK8 A0A1B6CTF4 A0A182HSW8 A0A182RGY2 A0A182XHY7 A0A182VWQ3 A0A084VAN0 A0A1B6BZ51 A0A2M4AW31 A0A0A1XI16 A0A2M4ASF9 A0A0K8WJB9 A0A2M4AUG5 M9PJR2 A0A034W6U3 W8BW02 Q59E36-3 A0A0R1EAT3 A0A182MCU5 A0A1A9VWZ2 A0A182YHK4 A0A1B6K8A3 A0A1L8DG32 A0A0Q5TJS9 A0A0P9AX75 A0A1B0ASU1 A0A1B0FLM9 A0A034W8H3 A0A1A9XYG8
Pubmed
28756777
19121390
22118469
26354079
18362917
19820115
+ More
25315136 28004739 24495485 25348373 25830018 26369729 26227816 17994087 17550304 10731132 12537572 12537569 15306652 17372656 18327897 17510324 12364791 20798317 24945155 26483478 21719571 24845553 26108605 24438588 12537568 12537573 12537574 16110336 17569856 17569867 25244985 18057021
25315136 28004739 24495485 25348373 25830018 26369729 26227816 17994087 17550304 10731132 12537572 12537569 15306652 17372656 18327897 17510324 12364791 20798317 24945155 26483478 21719571 24845553 26108605 24438588 12537568 12537573 12537574 16110336 17569856 17569867 25244985 18057021
EMBL
NWSH01000028
PCG80576.1
KZ150058
PZC74273.1
BABH01001791
AGBW02012095
+ More
OWR45688.1 PCG80575.1 PCG80577.1 ODYU01009812 SOQ54480.1 PCG80574.1 KQ459604 KPI92631.1 KQ971338 EFA01740.2 KA646202 AFP60831.1 GEZM01094222 JAV55798.1 KQ461196 KPJ06586.1 GAMC01005433 JAC01123.1 GAKP01008519 GAKP01008516 JAC50433.1 GBXI01015164 JAC99127.1 GDAI01002925 JAI14678.1 GANO01004410 JAB55461.1 GFDF01008583 JAV05501.1 JTDY01006763 KOB65710.1 CH954180 EDV46707.2 CM000162 EDX02130.2 AE014298 BT023817 AY060226 AY118507 CH902621 KPU81684.1 CH477208 EAT47760.1 CH940655 EDW66096.2 KRF82558.1 OUUW01000031 SPP89802.1 GEZM01094223 JAV55797.1 KYB27992.1 GAMC01005430 JAC01126.1 KQ414584 KOC70580.1 AJVK01015164 AJVK01015165 AAAB01008859 EAA08204.4 GL451103 EFN80005.1 GAMC01005431 JAC01125.1 GEDC01003680 JAS33618.1 NEVH01024529 PNF17016.1 GAPW01000532 JAC13066.1 KQ980204 KYN17250.1 JXUM01091414 KQ563913 KXJ73079.1 GL888525 EGI59843.1 GEDC01003481 JAS33817.1 AXCN02000175 KK852559 KDR21390.1 JRES01001467 KNC22656.1 GEDC01020546 JAS16752.1 APCN01000372 ATLV01004133 KE524190 KFB35024.1 GEDC01031008 JAS06290.1 GGFK01011659 MBW44980.1 GBXI01003690 JAD10602.1 GGFK01010379 MBW43700.1 GDHF01002228 GDHF01001091 JAI50086.1 JAI51223.1 GGFK01011041 MBW44362.1 AGB95546.1 GAKP01008518 JAC50434.1 GAMC01005432 JAC01124.1 KRK06579.1 KRK06582.1 AXCM01001329 GECU01000027 JAT07680.1 GFDF01008737 JAV05347.1 KQS30089.1 KQS30090.1 KPU81685.1 JXJN01002998 JXJN01002999 JXJN01003000 CCAG010001342 CCAG010001343 GAKP01008517 JAC50435.1
OWR45688.1 PCG80575.1 PCG80577.1 ODYU01009812 SOQ54480.1 PCG80574.1 KQ459604 KPI92631.1 KQ971338 EFA01740.2 KA646202 AFP60831.1 GEZM01094222 JAV55798.1 KQ461196 KPJ06586.1 GAMC01005433 JAC01123.1 GAKP01008519 GAKP01008516 JAC50433.1 GBXI01015164 JAC99127.1 GDAI01002925 JAI14678.1 GANO01004410 JAB55461.1 GFDF01008583 JAV05501.1 JTDY01006763 KOB65710.1 CH954180 EDV46707.2 CM000162 EDX02130.2 AE014298 BT023817 AY060226 AY118507 CH902621 KPU81684.1 CH477208 EAT47760.1 CH940655 EDW66096.2 KRF82558.1 OUUW01000031 SPP89802.1 GEZM01094223 JAV55797.1 KYB27992.1 GAMC01005430 JAC01126.1 KQ414584 KOC70580.1 AJVK01015164 AJVK01015165 AAAB01008859 EAA08204.4 GL451103 EFN80005.1 GAMC01005431 JAC01125.1 GEDC01003680 JAS33618.1 NEVH01024529 PNF17016.1 GAPW01000532 JAC13066.1 KQ980204 KYN17250.1 JXUM01091414 KQ563913 KXJ73079.1 GL888525 EGI59843.1 GEDC01003481 JAS33817.1 AXCN02000175 KK852559 KDR21390.1 JRES01001467 KNC22656.1 GEDC01020546 JAS16752.1 APCN01000372 ATLV01004133 KE524190 KFB35024.1 GEDC01031008 JAS06290.1 GGFK01011659 MBW44980.1 GBXI01003690 JAD10602.1 GGFK01010379 MBW43700.1 GDHF01002228 GDHF01001091 JAI50086.1 JAI51223.1 GGFK01011041 MBW44362.1 AGB95546.1 GAKP01008518 JAC50434.1 GAMC01005432 JAC01124.1 KRK06579.1 KRK06582.1 AXCM01001329 GECU01000027 JAT07680.1 GFDF01008737 JAV05347.1 KQS30089.1 KQS30090.1 KPU81685.1 JXJN01002998 JXJN01002999 JXJN01003000 CCAG010001342 CCAG010001343 GAKP01008517 JAC50435.1
Proteomes
UP000218220
UP000005204
UP000007151
UP000053268
UP000007266
UP000095301
+ More
UP000053240 UP000095300 UP000037510 UP000008711 UP000002282 UP000000803 UP000192223 UP000007801 UP000192221 UP000008820 UP000008792 UP000268350 UP000053825 UP000092462 UP000075902 UP000075884 UP000007062 UP000008237 UP000075881 UP000235965 UP000078492 UP000069940 UP000249989 UP000075880 UP000007755 UP000075886 UP000075903 UP000027135 UP000037069 UP000075840 UP000075900 UP000076407 UP000075920 UP000030765 UP000075883 UP000078200 UP000076408 UP000092460 UP000092444 UP000092443
UP000053240 UP000095300 UP000037510 UP000008711 UP000002282 UP000000803 UP000192223 UP000007801 UP000192221 UP000008820 UP000008792 UP000268350 UP000053825 UP000092462 UP000075902 UP000075884 UP000007062 UP000008237 UP000075881 UP000235965 UP000078492 UP000069940 UP000249989 UP000075880 UP000007755 UP000075886 UP000075903 UP000027135 UP000037069 UP000075840 UP000075900 UP000076407 UP000075920 UP000030765 UP000075883 UP000078200 UP000076408 UP000092460 UP000092444 UP000092443
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4K9A8
A0A2W1BR93
H9JFR7
A0A212EW08
A0A2A4K8H2
A0A2A4K8N8
+ More
A0A2H1WN35 A0A2A4K9J2 A0A194PGW2 D2A064 A0A1I8M1Q7 T1PBR0 A0A1Y1K5V9 A0A194QSS1 W8BQ39 A0A034W9G7 A0A0A1WL46 A0A0K8TKM1 A0A1I8NWR0 U5EQ84 A0A1L8DGE5 A0A0L7KRJ7 B3NVP2 B4PZH4 Q59E36 A0A1W4WQJ4 A0A1W4WPI9 A0A0N8P243 A0A1W4VUS2 Q17M84 B4MA37 A0A0Q9WP20 Q59E36-4 A0A3B0KAQ5 A0A1Y1KAM5 A0A139WJB2 W8BIL6 A0A0L7RI93 A0A1B0DG41 A0A182TW84 A0A182N535 Q7QC78 E2BWA1 W8CAT6 A0A1B6E6Q8 A0A182K191 A0A2J7PL06 A0A023EVK7 A0A195DWG3 A0A182GVN2 A0A1I8M1Q3 A0A182IM06 F4X132 A0A1B6E7D2 A0A182QVL8 A0A182VB33 A0A067RLA0 A0A0L0BRK8 A0A1B6CTF4 A0A182HSW8 A0A182RGY2 A0A182XHY7 A0A182VWQ3 A0A084VAN0 A0A1B6BZ51 A0A2M4AW31 A0A0A1XI16 A0A2M4ASF9 A0A0K8WJB9 A0A2M4AUG5 M9PJR2 A0A034W6U3 W8BW02 Q59E36-3 A0A0R1EAT3 A0A182MCU5 A0A1A9VWZ2 A0A182YHK4 A0A1B6K8A3 A0A1L8DG32 A0A0Q5TJS9 A0A0P9AX75 A0A1B0ASU1 A0A1B0FLM9 A0A034W8H3 A0A1A9XYG8
A0A2H1WN35 A0A2A4K9J2 A0A194PGW2 D2A064 A0A1I8M1Q7 T1PBR0 A0A1Y1K5V9 A0A194QSS1 W8BQ39 A0A034W9G7 A0A0A1WL46 A0A0K8TKM1 A0A1I8NWR0 U5EQ84 A0A1L8DGE5 A0A0L7KRJ7 B3NVP2 B4PZH4 Q59E36 A0A1W4WQJ4 A0A1W4WPI9 A0A0N8P243 A0A1W4VUS2 Q17M84 B4MA37 A0A0Q9WP20 Q59E36-4 A0A3B0KAQ5 A0A1Y1KAM5 A0A139WJB2 W8BIL6 A0A0L7RI93 A0A1B0DG41 A0A182TW84 A0A182N535 Q7QC78 E2BWA1 W8CAT6 A0A1B6E6Q8 A0A182K191 A0A2J7PL06 A0A023EVK7 A0A195DWG3 A0A182GVN2 A0A1I8M1Q3 A0A182IM06 F4X132 A0A1B6E7D2 A0A182QVL8 A0A182VB33 A0A067RLA0 A0A0L0BRK8 A0A1B6CTF4 A0A182HSW8 A0A182RGY2 A0A182XHY7 A0A182VWQ3 A0A084VAN0 A0A1B6BZ51 A0A2M4AW31 A0A0A1XI16 A0A2M4ASF9 A0A0K8WJB9 A0A2M4AUG5 M9PJR2 A0A034W6U3 W8BW02 Q59E36-3 A0A0R1EAT3 A0A182MCU5 A0A1A9VWZ2 A0A182YHK4 A0A1B6K8A3 A0A1L8DG32 A0A0Q5TJS9 A0A0P9AX75 A0A1B0ASU1 A0A1B0FLM9 A0A034W8H3 A0A1A9XYG8
PDB
4CZZ
E-value=3.48285e-53,
Score=525
Ontologies
GO
PANTHER
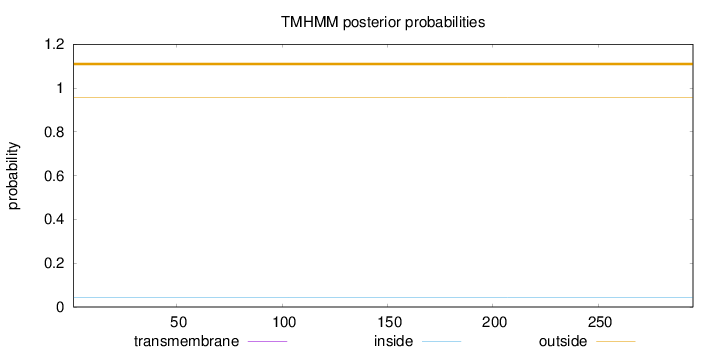
Topology
Subcellular location
Nucleus
Length:
295
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00148
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04408
outside
1 - 295
Population Genetic Test Statistics
Pi
190.021884
Theta
189.630901
Tajima's D
-0.383928
CLR
108.099019
CSRT
0.267686615669217
Interpretation
Uncertain
