Gene
KWMTBOMO10838
Pre Gene Modal
BGIBMGA008371
Annotation
PREDICTED:_protein_FAM177A1-like_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 1.545 Nuclear Reliability : 2.17
Sequence
CDS
ATGGCAAGTAACGAAAATAGTCAAATAAAGTTAAATATTCAACCAACTCGAGTATTACATTTTAGTGACGGAGTGGACGAAATAGTGGAAGAAGAGGAAGTGAATGAATTAAAAAACGAACCCAAGGAAGCGAACGTAGATCCTAAAACCCTTTCTTGGCTGCCTTGGCTTACGCATTATGCAAATACATCAGGCACTAAAGTGCTAAATGCAATTGATTATACCGGAGAGTCGCTTGCAAACTTCTTTGGCATAACAACACCAAAATATCAGCTTGAAATAGACGAGTATGAAAGATTAAAAGAAGAGAAAAAGAAAATGGAAGACGAGTCAGCTGGTTGGGTACCAAAGAATTCTGGTGGTGATATTCCTTTAGTGTTAAATGAGCCAGGAAAAGATGTAACAAGACAAGAAGCCTGA
Protein
MASNENSQIKLNIQPTRVLHFSDGVDEIVEEEEVNELKNEPKEANVDPKTLSWLPWLTHYANTSGTKVLNAIDYTGESLANFFGITTPKYQLEIDEYERLKEEKKKMEDESAGWVPKNSGGDIPLVLNEPGKDVTRQEA
Summary
Uniprot
H9JFS4
A0A2H1VP35
A0A2A4KA97
A0A2W1C127
A0A1E1WS35
S4PX70
+ More
A0A0L7LKG8 A0A194PCF3 A0A194QST0 A0A0T6B9U3 A0A2R5L9U5 A0A1B6JE93 A0A1B6IF65 A0A0M8ZU06 A0A1B6KA88 A0A1B6GHP3 A0A1B6FDY0 A0A026W8Z4 V9IDM9 A0A3L8DN02 A0A0P4XAC0 E2B8G7 A0A0P5FJ79 A0A0P6HWK9 A0A2P8XT93 A0A0P6I2S6 A0A146MHJ0 A0A0P5X5Q0 A0A0A9X1S1 F4X2K8 A0A0P4VMT6 A0A0P5YHU9 T1HI23 A0A2R2MPY3 E9GMG6 A0A195DRQ1 A0A0P6BCB0 A0A195FXP5 A0A0L7R065 A0A158NVA3 A0A195BS76 A0A151IBE1 E9J9R3 A0A151WFT5 A0A2A3EHJ4 A0A0K8RRI5 N6TNU3 N6ULF1 N6TFF0 A0A067QZP6 A0A226EXW8 A0A131Y9I0 A0A1J1IBI1 A0A1Y1N1A3 A0A2C9LAZ5 D2A1B8 A0A0L8GT04 A0A1W0WSC1 L7MBJ9 L7MB73 A0A170Z012 A0A224YDT8 A0A2R7W1Z0 A0A131XFF1 A0A131YXU1 A0A2M4CR74 A0A2M4CR80 A0A2M4ATT3 W5JUY5 A0A2M4ATZ6 A0A2M4AUA5 A0A2M4CRA2 A0A182FT00 A0A2M3ZJY7 A0A2M4C0V5 A0A2M4C0W7 B0WZK3 A0A0K8TPW3 U4UFH8 A0A1W4WIP9 A0A1Q3FFY9 A0A2J7PEI8 T1E822 A0A023EJ80 Q17H54
A0A0L7LKG8 A0A194PCF3 A0A194QST0 A0A0T6B9U3 A0A2R5L9U5 A0A1B6JE93 A0A1B6IF65 A0A0M8ZU06 A0A1B6KA88 A0A1B6GHP3 A0A1B6FDY0 A0A026W8Z4 V9IDM9 A0A3L8DN02 A0A0P4XAC0 E2B8G7 A0A0P5FJ79 A0A0P6HWK9 A0A2P8XT93 A0A0P6I2S6 A0A146MHJ0 A0A0P5X5Q0 A0A0A9X1S1 F4X2K8 A0A0P4VMT6 A0A0P5YHU9 T1HI23 A0A2R2MPY3 E9GMG6 A0A195DRQ1 A0A0P6BCB0 A0A195FXP5 A0A0L7R065 A0A158NVA3 A0A195BS76 A0A151IBE1 E9J9R3 A0A151WFT5 A0A2A3EHJ4 A0A0K8RRI5 N6TNU3 N6ULF1 N6TFF0 A0A067QZP6 A0A226EXW8 A0A131Y9I0 A0A1J1IBI1 A0A1Y1N1A3 A0A2C9LAZ5 D2A1B8 A0A0L8GT04 A0A1W0WSC1 L7MBJ9 L7MB73 A0A170Z012 A0A224YDT8 A0A2R7W1Z0 A0A131XFF1 A0A131YXU1 A0A2M4CR74 A0A2M4CR80 A0A2M4ATT3 W5JUY5 A0A2M4ATZ6 A0A2M4AUA5 A0A2M4CRA2 A0A182FT00 A0A2M3ZJY7 A0A2M4C0V5 A0A2M4C0W7 B0WZK3 A0A0K8TPW3 U4UFH8 A0A1W4WIP9 A0A1Q3FFY9 A0A2J7PEI8 T1E822 A0A023EJ80 Q17H54
Pubmed
EMBL
BABH01001765
ODYU01003357
SOQ42014.1
NWSH01000028
PCG80572.1
KZ149919
+ More
PZC77783.1 GDQN01001327 JAT89727.1 GAIX01005553 JAA87007.1 JTDY01000851 KOB75671.1 KQ459606 KPI90882.1 KQ461196 KPJ06596.1 LJIG01002818 KRT84122.1 GGLE01002099 MBY06225.1 GECU01010156 JAS97550.1 GECU01022166 JAS85540.1 KQ435878 KOX69966.1 GEBQ01031863 JAT08114.1 GECZ01007823 JAS61946.1 GECZ01021445 JAS48324.1 KK107324 EZA52530.1 JR040563 AEY59185.1 QOIP01000006 RLU21656.1 GDIP01245964 GDIP01245572 LRGB01001570 JAI77437.1 KZS11731.1 GL446332 EFN88001.1 GDIQ01256282 JAJ95442.1 GDIQ01028771 JAN65966.1 PYGN01001381 PSN35238.1 GDIQ01010530 JAN84207.1 GDHC01006571 GDHC01000479 JAQ12058.1 JAQ18150.1 GDIP01077064 JAM26651.1 GBHO01030871 GBHO01030870 GBRD01016643 GBRD01016642 GBRD01016641 GBRD01016640 GBRD01002342 GBRD01002340 GBRD01002339 GBRD01002338 GBRD01000233 JAG12733.1 JAG12734.1 JAG49184.1 GL888584 EGI59333.1 GDKW01003554 JAI53041.1 GDIP01057651 JAM46064.1 ACPB03012932 GL732552 EFX79394.1 KQ980530 KYN15605.1 GDIP01017276 JAM86439.1 KQ981193 KYN45201.1 KQ414672 KOC64233.1 ADTU01027051 KQ976417 KYM89882.1 KQ978106 KYM96973.1 GL769399 EFZ10458.1 KQ983203 KYQ46693.1 KZ288246 PBC31187.1 GADI01000106 JAA73702.1 APGK01011739 APGK01015630 KB739628 KB738560 ENN82189.1 ENN82554.1 APGK01011729 APGK01011730 KB738559 ENN82555.1 APGK01029891 KB740752 ENN79069.1 KK853150 KDR10606.1 LNIX01000001 OXA62028.1 GEFM01000598 JAP75198.1 CVRI01000046 CRK97098.1 GEZM01017461 GEZM01017460 JAV90660.1 KQ971338 EFA01553.1 KQ420550 KOF79959.1 MTYJ01000053 OQV18095.1 GACK01003423 JAA61611.1 GACK01003533 JAA61501.1 GEMB01002767 JAS00430.1 GFPF01004720 MAA15866.1 KK854230 PTY13448.1 GEFH01002688 JAP65893.1 GEDV01005262 JAP83295.1 GGFL01003654 MBW67832.1 GGFL01003656 MBW67834.1 GGFK01010894 MBW44215.1 ADMH02000418 ETN66584.1 GGFK01010922 MBW44243.1 GGFK01011032 MBW44353.1 GGFL01003655 MBW67833.1 GGFM01008029 MBW28780.1 GGFJ01009799 MBW58940.1 GGFJ01009798 MBW58939.1 DS232209 EDS37517.1 GDAI01001161 JAI16442.1 KB632166 ERL89326.1 GFDL01008575 JAV26470.1 NEVH01026094 PNF14754.1 GAMD01002582 JAA99008.1 GAPW01004719 JAC08879.1 CH477253 EAT45959.1
PZC77783.1 GDQN01001327 JAT89727.1 GAIX01005553 JAA87007.1 JTDY01000851 KOB75671.1 KQ459606 KPI90882.1 KQ461196 KPJ06596.1 LJIG01002818 KRT84122.1 GGLE01002099 MBY06225.1 GECU01010156 JAS97550.1 GECU01022166 JAS85540.1 KQ435878 KOX69966.1 GEBQ01031863 JAT08114.1 GECZ01007823 JAS61946.1 GECZ01021445 JAS48324.1 KK107324 EZA52530.1 JR040563 AEY59185.1 QOIP01000006 RLU21656.1 GDIP01245964 GDIP01245572 LRGB01001570 JAI77437.1 KZS11731.1 GL446332 EFN88001.1 GDIQ01256282 JAJ95442.1 GDIQ01028771 JAN65966.1 PYGN01001381 PSN35238.1 GDIQ01010530 JAN84207.1 GDHC01006571 GDHC01000479 JAQ12058.1 JAQ18150.1 GDIP01077064 JAM26651.1 GBHO01030871 GBHO01030870 GBRD01016643 GBRD01016642 GBRD01016641 GBRD01016640 GBRD01002342 GBRD01002340 GBRD01002339 GBRD01002338 GBRD01000233 JAG12733.1 JAG12734.1 JAG49184.1 GL888584 EGI59333.1 GDKW01003554 JAI53041.1 GDIP01057651 JAM46064.1 ACPB03012932 GL732552 EFX79394.1 KQ980530 KYN15605.1 GDIP01017276 JAM86439.1 KQ981193 KYN45201.1 KQ414672 KOC64233.1 ADTU01027051 KQ976417 KYM89882.1 KQ978106 KYM96973.1 GL769399 EFZ10458.1 KQ983203 KYQ46693.1 KZ288246 PBC31187.1 GADI01000106 JAA73702.1 APGK01011739 APGK01015630 KB739628 KB738560 ENN82189.1 ENN82554.1 APGK01011729 APGK01011730 KB738559 ENN82555.1 APGK01029891 KB740752 ENN79069.1 KK853150 KDR10606.1 LNIX01000001 OXA62028.1 GEFM01000598 JAP75198.1 CVRI01000046 CRK97098.1 GEZM01017461 GEZM01017460 JAV90660.1 KQ971338 EFA01553.1 KQ420550 KOF79959.1 MTYJ01000053 OQV18095.1 GACK01003423 JAA61611.1 GACK01003533 JAA61501.1 GEMB01002767 JAS00430.1 GFPF01004720 MAA15866.1 KK854230 PTY13448.1 GEFH01002688 JAP65893.1 GEDV01005262 JAP83295.1 GGFL01003654 MBW67832.1 GGFL01003656 MBW67834.1 GGFK01010894 MBW44215.1 ADMH02000418 ETN66584.1 GGFK01010922 MBW44243.1 GGFK01011032 MBW44353.1 GGFL01003655 MBW67833.1 GGFM01008029 MBW28780.1 GGFJ01009799 MBW58940.1 GGFJ01009798 MBW58939.1 DS232209 EDS37517.1 GDAI01001161 JAI16442.1 KB632166 ERL89326.1 GFDL01008575 JAV26470.1 NEVH01026094 PNF14754.1 GAMD01002582 JAA99008.1 GAPW01004719 JAC08879.1 CH477253 EAT45959.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000053105
+ More
UP000053097 UP000279307 UP000076858 UP000008237 UP000245037 UP000007755 UP000015103 UP000085678 UP000000305 UP000078492 UP000078541 UP000053825 UP000005205 UP000078540 UP000078542 UP000075809 UP000242457 UP000019118 UP000027135 UP000198287 UP000183832 UP000076420 UP000007266 UP000053454 UP000000673 UP000069272 UP000002320 UP000030742 UP000192223 UP000235965 UP000008820
UP000053097 UP000279307 UP000076858 UP000008237 UP000245037 UP000007755 UP000015103 UP000085678 UP000000305 UP000078492 UP000078541 UP000053825 UP000005205 UP000078540 UP000078542 UP000075809 UP000242457 UP000019118 UP000027135 UP000198287 UP000183832 UP000076420 UP000007266 UP000053454 UP000000673 UP000069272 UP000002320 UP000030742 UP000192223 UP000235965 UP000008820
PRIDE
Pfam
PF14774 FAM177
ProteinModelPortal
H9JFS4
A0A2H1VP35
A0A2A4KA97
A0A2W1C127
A0A1E1WS35
S4PX70
+ More
A0A0L7LKG8 A0A194PCF3 A0A194QST0 A0A0T6B9U3 A0A2R5L9U5 A0A1B6JE93 A0A1B6IF65 A0A0M8ZU06 A0A1B6KA88 A0A1B6GHP3 A0A1B6FDY0 A0A026W8Z4 V9IDM9 A0A3L8DN02 A0A0P4XAC0 E2B8G7 A0A0P5FJ79 A0A0P6HWK9 A0A2P8XT93 A0A0P6I2S6 A0A146MHJ0 A0A0P5X5Q0 A0A0A9X1S1 F4X2K8 A0A0P4VMT6 A0A0P5YHU9 T1HI23 A0A2R2MPY3 E9GMG6 A0A195DRQ1 A0A0P6BCB0 A0A195FXP5 A0A0L7R065 A0A158NVA3 A0A195BS76 A0A151IBE1 E9J9R3 A0A151WFT5 A0A2A3EHJ4 A0A0K8RRI5 N6TNU3 N6ULF1 N6TFF0 A0A067QZP6 A0A226EXW8 A0A131Y9I0 A0A1J1IBI1 A0A1Y1N1A3 A0A2C9LAZ5 D2A1B8 A0A0L8GT04 A0A1W0WSC1 L7MBJ9 L7MB73 A0A170Z012 A0A224YDT8 A0A2R7W1Z0 A0A131XFF1 A0A131YXU1 A0A2M4CR74 A0A2M4CR80 A0A2M4ATT3 W5JUY5 A0A2M4ATZ6 A0A2M4AUA5 A0A2M4CRA2 A0A182FT00 A0A2M3ZJY7 A0A2M4C0V5 A0A2M4C0W7 B0WZK3 A0A0K8TPW3 U4UFH8 A0A1W4WIP9 A0A1Q3FFY9 A0A2J7PEI8 T1E822 A0A023EJ80 Q17H54
A0A0L7LKG8 A0A194PCF3 A0A194QST0 A0A0T6B9U3 A0A2R5L9U5 A0A1B6JE93 A0A1B6IF65 A0A0M8ZU06 A0A1B6KA88 A0A1B6GHP3 A0A1B6FDY0 A0A026W8Z4 V9IDM9 A0A3L8DN02 A0A0P4XAC0 E2B8G7 A0A0P5FJ79 A0A0P6HWK9 A0A2P8XT93 A0A0P6I2S6 A0A146MHJ0 A0A0P5X5Q0 A0A0A9X1S1 F4X2K8 A0A0P4VMT6 A0A0P5YHU9 T1HI23 A0A2R2MPY3 E9GMG6 A0A195DRQ1 A0A0P6BCB0 A0A195FXP5 A0A0L7R065 A0A158NVA3 A0A195BS76 A0A151IBE1 E9J9R3 A0A151WFT5 A0A2A3EHJ4 A0A0K8RRI5 N6TNU3 N6ULF1 N6TFF0 A0A067QZP6 A0A226EXW8 A0A131Y9I0 A0A1J1IBI1 A0A1Y1N1A3 A0A2C9LAZ5 D2A1B8 A0A0L8GT04 A0A1W0WSC1 L7MBJ9 L7MB73 A0A170Z012 A0A224YDT8 A0A2R7W1Z0 A0A131XFF1 A0A131YXU1 A0A2M4CR74 A0A2M4CR80 A0A2M4ATT3 W5JUY5 A0A2M4ATZ6 A0A2M4AUA5 A0A2M4CRA2 A0A182FT00 A0A2M3ZJY7 A0A2M4C0V5 A0A2M4C0W7 B0WZK3 A0A0K8TPW3 U4UFH8 A0A1W4WIP9 A0A1Q3FFY9 A0A2J7PEI8 T1E822 A0A023EJ80 Q17H54
Ontologies
GO
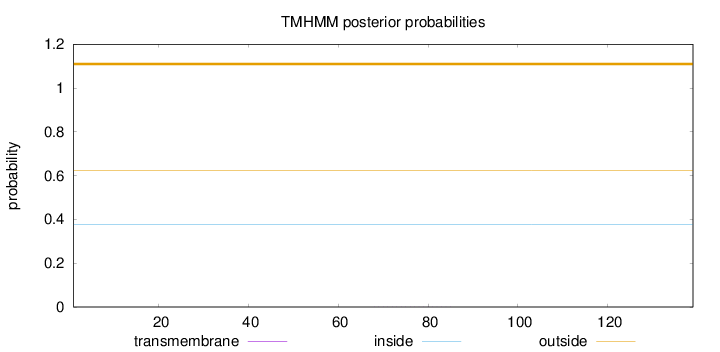
Topology
Length:
139
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00444
Exp number, first 60 AAs:
0.00044
Total prob of N-in:
0.37569
outside
1 - 139
Population Genetic Test Statistics
Pi
228.533769
Theta
178.571118
Tajima's D
0.90349
CLR
65.922464
CSRT
0.633918304084796
Interpretation
Uncertain
