Gene
KWMTBOMO10834
Pre Gene Modal
BGIBMGA008375
Annotation
PREDICTED:_uncharacterized_protein_LOC101741673?_partial_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.973
Sequence
CDS
ATGAGCCCACTGAGTCTCCTTAACGAGTACTATATACCGGACGACCCGGAAAAACAAGAGACGATCCTGCGGAAGCTATCAGCCTACTGCGGCAAGTCTGTCCTCAACTCAACGACAGAAGATACAGAGGCTCCATACAAATGGGACTTCTACCATTCATTCTTCTTCTCTTATACGGTCGTCAGTACTATTGGTTACGGTAACTTGTCCCCGACGACACATTTATCGCGTATTCTTATGATATTCTACGGTCTCTTTGGCATACCCATCAACGGTATTCTACTTGCAAATCTCGGTGAATATTTCGGTCTCCAGCTAATATCAGTATACCGTAAATATAAGAAGAAAAATGAAAAACGAGCCGATAACTTGGACTTTTTCTTCAATAACCTTGGCATGCTCGGGCAGATTTTTCTTTATCTGGTCCCCGGATTTCTGTTCTTCATATTCTTGCCAGCTTGTATTTTCGTCGTGTTTGAAGGCTGGGATTATGTTGCGGCCATATATTACGCCTTTGTCACTCTCACGACGATTGGATTTGGTGACTTGGTTGCAGGAACTGTCAACAATGGTTTCAAGTGGGGTTACTTTTTTACCTACCAGATATTCCTGATAGTCTGGATCACATTTGGACTTGGCTACATCGTGATGTTACTAGGTTTCATTACTAGTGGCATGAGATCAGAACGCATACATAGAATAGAACAGAAGTTTGCCTATCAATTTAAGGCCACGCAGAACAAGATTCTACAAGGATTCACCAAAGACATTTCCGCTCTTAGGAAAATCATAAACGAAGCCAATTTGATCAAAATTAAGCCAATATATGTCGATACAACACCACACATTACGAGAAGTGCCAGCTGTCCAACGTTCACTTTTGACGTTGAACCAAGAGATCCAATCGTGAAACGAAAACGAGCAAATTCTGAAAATATTCAACTTTCTGCCAAAGATATGCTACGAATACAATCAGACACTGATTTACTAGGAATAGACAAAGAAAAGACATTCACAGCTTCTGCTCTCGTAAAACCAGCGGAACTCTTAGCTCGAGTAGTAAACGTACTCGGAGGCTTTGAATCACCTCAGCAAAAGTCTGGTATCAACTTGTTTGATGATGATCATATTTTAGCAAATGAAAAACCACCATTATTTAGCATAGGCCAAGATCTCATTACTAACTCACCAAAGAGAGCTAGGGCGCTCAGCGTGGCTGTACCGAATTACAGAAAAGAATCTTTGGCCACGAAACTAGATCATGATTTCACATGGACTAGCGGAGAATCAGGAGATAAGCTTAGACGAGAAGCTAATATGAGAAGTGGGAAATTATCACTGCCCGCTAAAGTCCCTCATTCAGATTGCTCTCAACCGCAAAGTATTATCTCAAAATTCTTCTTCAGAAAACAAAGTTCAACTGATAATCCGGCCGAATATATCAAAAGCTCTATGAGGGATCGGGTTAACAGTGCACAAGGCAGCACTGAAAGCATTAAAGACAATGGAAAACCTAGGCGTGGAAGTATATTTCCTTCATTTATATTCGGATCGTCTGAAGGAAAAAACGAAGCTGATGCATACAAAGAAAAGACAAAACGAGGGCGCCATAGTATCTTTCCCACTTTTGATTTCCATCGTAGCGAAGAAGATTCTGCAAACGCTTACCTAGAAAAGACTAAGCGTGGTCGTCACAGCATTTTTCCCACATTTGATTTCTCGGAAGACGAAGACTTTGCCGCAATGCGAGCTTACCAACAGAAACCTAAGCGGCACAGTATATTTCCTACTTTTGATTTTAGTGAAGATGAAAATGCATCTGCTGCTAAAGCCTATAAAGAAAAAACGAAACAAGGAAGACGTAGTATTTTCCCTACATTCGATTTTAGCGATCCTGACGATGATATGGCTGAGCGTTATAAAGAAAATACAAGGCGTGGTAGACGCAGCATTTTCCCATCATTCAATTTCAGCGGTCTTACAAGTAATGATGAGGATGATACAGTAGACGAAGATGAAGTGCAGAGATATCAAAACCGTACAAGGAAAGGTCGATTAAGTCTGTTTCCTACTTTCGGCAAGTCCAAGAAGAACGACGACGATAACTTGCCTGAAAGCTCTGACGAACTTGTAGATTATAAAAACAAGACCCGCCGCGGTCGCAGTAGTCTGTTTCCTTCTTTCGGAAAGGAACGTAAAGACAGTGCACAAACAGATGATCCCAGTGATCTCGAAACTAGTATTCGACAATACCAAAAGCAAAGACTGGGACGGAATAGTGCTTTCGACGGAGATGATTCCAGGAATTCTACTCCAGATAGAAGAAACAGTATTTTCTCTAGTGAAATAGAAAATTATAGAAGTAAAACAAGAGGCGGCCGTCCTAGTCTTTTTGACCCAGATGTAAATATTTCAAACTTACCACAATCAGAGCAAGTGGAAGTTTTAGAGCATACTAGTGTTGCTGATTTACTGAGAGCTCTCGCAGCATTAGAGGCAGCTGGTCAAAATCCTTACAGCTCTAATACTGGTGAGCCATCAGGCTTATTAGAAATGATATCTGAACCCTTGTCTGACACATCTAGAAGACGAGGCTCCTTGAGACATGATCTTCAGTCACAAACTCCTCCTGGTTGCGTAGAAGACTGTACTGGTCCACGCCGTCGTCGAATATCAGCTAGAGCGGCTGCTCCTCAATTTACTCCAACGCTGGCAACTGTTGTTGCTGGTACGGAATTACAGTTAAGTACCGCAACAGCTGCTAGAGAAAATCGCATGTCCTTGTTAACTCAGCCTCCACCCCCCTACTCTGAAACTCCTAGTGAAGAAGAGAACAAGCCTCGTCTTAGAAGATATTCCCCTGCGCCTGTACCTAGTGATTCTGGAAGGTCTGCAGGTCCTGTACCGAGACTGTTTTCAAGGATGCGCAAAGAAAGCGCCCCCAATGCGGAGGACGGGGACTCGAGAAGAGGTAGCCTAACCGATATAGTCATTGATAGTTTTAAAGATAAAACGTGA
Protein
MSPLSLLNEYYIPDDPEKQETILRKLSAYCGKSVLNSTTEDTEAPYKWDFYHSFFFSYTVVSTIGYGNLSPTTHLSRILMIFYGLFGIPINGILLANLGEYFGLQLISVYRKYKKKNEKRADNLDFFFNNLGMLGQIFLYLVPGFLFFIFLPACIFVVFEGWDYVAAIYYAFVTLTTIGFGDLVAGTVNNGFKWGYFFTYQIFLIVWITFGLGYIVMLLGFITSGMRSERIHRIEQKFAYQFKATQNKILQGFTKDISALRKIINEANLIKIKPIYVDTTPHITRSASCPTFTFDVEPRDPIVKRKRANSENIQLSAKDMLRIQSDTDLLGIDKEKTFTASALVKPAELLARVVNVLGGFESPQQKSGINLFDDDHILANEKPPLFSIGQDLITNSPKRARALSVAVPNYRKESLATKLDHDFTWTSGESGDKLRREANMRSGKLSLPAKVPHSDCSQPQSIISKFFFRKQSSTDNPAEYIKSSMRDRVNSAQGSTESIKDNGKPRRGSIFPSFIFGSSEGKNEADAYKEKTKRGRHSIFPTFDFHRSEEDSANAYLEKTKRGRHSIFPTFDFSEDEDFAAMRAYQQKPKRHSIFPTFDFSEDENASAAKAYKEKTKQGRRSIFPTFDFSDPDDDMAERYKENTRRGRRSIFPSFNFSGLTSNDEDDTVDEDEVQRYQNRTRKGRLSLFPTFGKSKKNDDDNLPESSDELVDYKNKTRRGRSSLFPSFGKERKDSAQTDDPSDLETSIRQYQKQRLGRNSAFDGDDSRNSTPDRRNSIFSSEIENYRSKTRGGRPSLFDPDVNISNLPQSEQVEVLEHTSVADLLRALAALEAAGQNPYSSNTGEPSGLLEMISEPLSDTSRRRGSLRHDLQSQTPPGCVEDCTGPRRRRISARAAAPQFTPTLATVVAGTELQLSTATAARENRMSLLTQPPPPYSETPSEEENKPRLRRYSPAPVPSDSGRSAGPVPRLFSRMRKESAPNAEDGDSRRGSLTDIVIDSFKDKT
Summary
Similarity
Belongs to the two pore domain potassium channel (TC 1.A.1.8) family.
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF07885 Ion_trans_2
CDD
ProteinModelPortal
PDB
4XDL
E-value=1.971e-28,
Score=317
Ontologies
GO
Topology
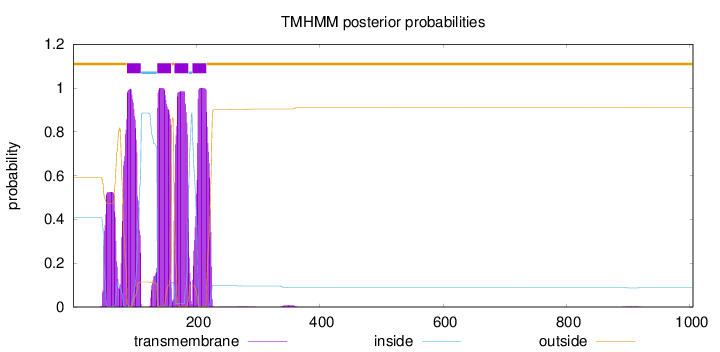
Length:
1005
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
103.30414
Exp number, first 60 AAs:
5.22502
Total prob of N-in:
0.40980
outside
1 - 87
TMhelix
88 - 110
inside
111 - 136
TMhelix
137 - 159
outside
160 - 164
TMhelix
165 - 187
inside
188 - 193
TMhelix
194 - 216
outside
217 - 1005
Population Genetic Test Statistics
Pi
234.942166
Theta
210.881456
Tajima's D
0.299516
CLR
0.037795
CSRT
0.454077296135193
Interpretation
Uncertain
