Gene
KWMTBOMO10828
Annotation
PREDICTED:_dual_specificity_tyrosine-phosphorylation-regulated_kinase_1A_isoform_X4_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 1.54 Nuclear Reliability : 2.081
Sequence
CDS
ATGGCAGCGTGGAACCGCCATCACCACTTCAACATGCTCATGGATGAAAATGCCAAAAACAAATTGGGCGTTGTCGGCGGCGGCATGCCAGAAGCCGCCGGTGGCGAGAGGCGGCACGTGCCGCTGTATGGCCGGCTGGTGGCTGAAGAACAGCTCGCGTCCATGAGCGCGCCTTCAGACATTCAGGCTATGCAAGCCAGAATACCACATCACTTTCGGGATCCTTCGACAGCGCCGCTACGGAAACTTAGCGTCGACCTCATTAAAACTTACAAACACATAAACGAGGAGACTTCCAAGACCGAACGTAAGCCCTGTCACTGA
Protein
MAAWNRHHHFNMLMDENAKNKLGVVGGGMPEAAGGERRHVPLYGRLVAEEQLASMSAPSDIQAMQARIPHHFRDPSTAPLRKLSVDLIKTYKHINEETSKTERKPCH
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
A0A2A4JRZ7
A0A2A4JRT9
A0A2W1BRX5
A0A2A4JS00
A0A2H1V2M1
A0A2A4JQA8
+ More
A0A212EI95 A0A146LQB0 A0A194QST9 A0A194PD08 A0A1Y1N919 A0A1W4WIK9 A0A1W4W6X5 A0A2P8Z2I9 D2A1V3 A0A1B6F8E6 N6U707 A0A1B6J3P2 A0A336LRQ8 A0A1W4WIC2 A0A182FRA0 V5GL93 A0A1B6KQA7 U4UN98 A0A1J1ID49 A0A1W4WHA8 A0A1A9W8U5 A0A1B6C8U4 A0A336K4G2 A0A1I8NPB4 A0A0A1WP88 A0A034V8T2 W8BKM4 A0A1I8NPB8 A0A0K8UWN4 A0A2M4CK37 A0A0K8W195 A0A2M3ZFF0 A0A1Q3EU33 A0A2M4BBK7 A0A0C9QZM2 A0A2M4A1I7 A0A3L8DEM8 A0A2M4B975 A0A0A1XLU7 A0A2M4CS68 A0A2M4CRQ0 A0A2M4CRW5 A0A2M4A0T0 A0A0C9Q5A0 A0A0A9VXE5 Q17FM4 A0A0K8TG95 A0A1S4G6S6 A0A0A9VT26 U5EZJ5 A0A195BVZ3 A0A034VDQ7 J9JQJ9 K7IR41 T1D614 A0A2H8TM44 T1DJG0 A0A2S2Q8F2 A0A2A3E0D4 A0A2S2NHL5 A0A2S2PX76 A0A0Q9WTY8 F5HMZ7 A0A1Y9IUX8 A0A1B0CIJ9 F5HMZ6 B4NCQ7 F5HMZ8
A0A212EI95 A0A146LQB0 A0A194QST9 A0A194PD08 A0A1Y1N919 A0A1W4WIK9 A0A1W4W6X5 A0A2P8Z2I9 D2A1V3 A0A1B6F8E6 N6U707 A0A1B6J3P2 A0A336LRQ8 A0A1W4WIC2 A0A182FRA0 V5GL93 A0A1B6KQA7 U4UN98 A0A1J1ID49 A0A1W4WHA8 A0A1A9W8U5 A0A1B6C8U4 A0A336K4G2 A0A1I8NPB4 A0A0A1WP88 A0A034V8T2 W8BKM4 A0A1I8NPB8 A0A0K8UWN4 A0A2M4CK37 A0A0K8W195 A0A2M3ZFF0 A0A1Q3EU33 A0A2M4BBK7 A0A0C9QZM2 A0A2M4A1I7 A0A3L8DEM8 A0A2M4B975 A0A0A1XLU7 A0A2M4CS68 A0A2M4CRQ0 A0A2M4CRW5 A0A2M4A0T0 A0A0C9Q5A0 A0A0A9VXE5 Q17FM4 A0A0K8TG95 A0A1S4G6S6 A0A0A9VT26 U5EZJ5 A0A195BVZ3 A0A034VDQ7 J9JQJ9 K7IR41 T1D614 A0A2H8TM44 T1DJG0 A0A2S2Q8F2 A0A2A3E0D4 A0A2S2NHL5 A0A2S2PX76 A0A0Q9WTY8 F5HMZ7 A0A1Y9IUX8 A0A1B0CIJ9 F5HMZ6 B4NCQ7 F5HMZ8
Pubmed
EMBL
NWSH01000790
PCG74180.1
PCG74183.1
KZ149919
PZC77798.1
PCG74182.1
+ More
ODYU01000386 SOQ35083.1 PCG74181.1 AGBW02014677 OWR41212.1 GDHC01009663 JAQ08966.1 KQ461196 KPJ06606.1 KQ459606 KPI90893.1 GEZM01013108 JAV92746.1 PYGN01000224 PSN50709.1 KQ971338 EFA02082.2 GECZ01023257 JAS46512.1 APGK01046974 KB741073 ENN74372.1 GECU01013976 JAS93730.1 UFQS01000056 UFQT01000056 SSW98721.1 SSX19107.1 GALX01003602 JAB64864.1 GEBQ01026370 JAT13607.1 KB632433 ERL95564.1 CVRI01000047 CRK97476.1 GEDC01027623 JAS09675.1 SSW98722.1 SSX19108.1 GBXI01013458 GBXI01006865 JAD00834.1 JAD07427.1 GAKP01019276 GAKP01019271 GAKP01019266 JAC39681.1 GAMC01012727 GAMC01012724 JAB93831.1 GDHF01021220 JAI31094.1 GGFL01001544 MBW65722.1 GDHF01007401 JAI44913.1 GGFM01006535 MBW27286.1 GFDL01016221 JAV18824.1 GGFJ01001276 MBW50417.1 GBYB01009229 JAG78996.1 GGFK01001336 MBW34657.1 QOIP01000009 RLU18854.1 GGFJ01000442 MBW49583.1 GBXI01002360 JAD11932.1 GGFL01003833 MBW68011.1 GGFL01003836 MBW68014.1 GGFL01003837 MBW68015.1 GGFK01001048 MBW34369.1 GBYB01009228 JAG78995.1 GBHO01044731 GBRD01001396 GDHC01018712 JAF98872.1 JAG64425.1 JAP99916.1 CH477270 EAT45319.1 GBRD01001395 JAG64426.1 GBHO01044730 GBRD01001398 GDHC01000811 JAF98873.1 JAG64423.1 JAQ17818.1 GANO01000682 JAB59189.1 KQ976398 KYM92804.1 GAKP01019281 GAKP01019279 GAKP01019274 JAC39673.1 ABLF02033064 GALA01000283 JAA94569.1 GFXV01002543 MBW14348.1 GALA01000286 JAA94566.1 GGMS01004289 MBY73492.1 KZ288488 PBC25225.1 GGMR01004065 MBY16684.1 GGMS01000790 MBY69993.1 CH964239 KRF99647.1 AAAB01008987 EGK97379.1 AJWK01013477 AJWK01013478 EGK97377.1 EDW82616.1 EGK97378.1
ODYU01000386 SOQ35083.1 PCG74181.1 AGBW02014677 OWR41212.1 GDHC01009663 JAQ08966.1 KQ461196 KPJ06606.1 KQ459606 KPI90893.1 GEZM01013108 JAV92746.1 PYGN01000224 PSN50709.1 KQ971338 EFA02082.2 GECZ01023257 JAS46512.1 APGK01046974 KB741073 ENN74372.1 GECU01013976 JAS93730.1 UFQS01000056 UFQT01000056 SSW98721.1 SSX19107.1 GALX01003602 JAB64864.1 GEBQ01026370 JAT13607.1 KB632433 ERL95564.1 CVRI01000047 CRK97476.1 GEDC01027623 JAS09675.1 SSW98722.1 SSX19108.1 GBXI01013458 GBXI01006865 JAD00834.1 JAD07427.1 GAKP01019276 GAKP01019271 GAKP01019266 JAC39681.1 GAMC01012727 GAMC01012724 JAB93831.1 GDHF01021220 JAI31094.1 GGFL01001544 MBW65722.1 GDHF01007401 JAI44913.1 GGFM01006535 MBW27286.1 GFDL01016221 JAV18824.1 GGFJ01001276 MBW50417.1 GBYB01009229 JAG78996.1 GGFK01001336 MBW34657.1 QOIP01000009 RLU18854.1 GGFJ01000442 MBW49583.1 GBXI01002360 JAD11932.1 GGFL01003833 MBW68011.1 GGFL01003836 MBW68014.1 GGFL01003837 MBW68015.1 GGFK01001048 MBW34369.1 GBYB01009228 JAG78995.1 GBHO01044731 GBRD01001396 GDHC01018712 JAF98872.1 JAG64425.1 JAP99916.1 CH477270 EAT45319.1 GBRD01001395 JAG64426.1 GBHO01044730 GBRD01001398 GDHC01000811 JAF98873.1 JAG64423.1 JAQ17818.1 GANO01000682 JAB59189.1 KQ976398 KYM92804.1 GAKP01019281 GAKP01019279 GAKP01019274 JAC39673.1 ABLF02033064 GALA01000283 JAA94569.1 GFXV01002543 MBW14348.1 GALA01000286 JAA94566.1 GGMS01004289 MBY73492.1 KZ288488 PBC25225.1 GGMR01004065 MBY16684.1 GGMS01000790 MBY69993.1 CH964239 KRF99647.1 AAAB01008987 EGK97379.1 AJWK01013477 AJWK01013478 EGK97377.1 EDW82616.1 EGK97378.1
Proteomes
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A2A4JRZ7
A0A2A4JRT9
A0A2W1BRX5
A0A2A4JS00
A0A2H1V2M1
A0A2A4JQA8
+ More
A0A212EI95 A0A146LQB0 A0A194QST9 A0A194PD08 A0A1Y1N919 A0A1W4WIK9 A0A1W4W6X5 A0A2P8Z2I9 D2A1V3 A0A1B6F8E6 N6U707 A0A1B6J3P2 A0A336LRQ8 A0A1W4WIC2 A0A182FRA0 V5GL93 A0A1B6KQA7 U4UN98 A0A1J1ID49 A0A1W4WHA8 A0A1A9W8U5 A0A1B6C8U4 A0A336K4G2 A0A1I8NPB4 A0A0A1WP88 A0A034V8T2 W8BKM4 A0A1I8NPB8 A0A0K8UWN4 A0A2M4CK37 A0A0K8W195 A0A2M3ZFF0 A0A1Q3EU33 A0A2M4BBK7 A0A0C9QZM2 A0A2M4A1I7 A0A3L8DEM8 A0A2M4B975 A0A0A1XLU7 A0A2M4CS68 A0A2M4CRQ0 A0A2M4CRW5 A0A2M4A0T0 A0A0C9Q5A0 A0A0A9VXE5 Q17FM4 A0A0K8TG95 A0A1S4G6S6 A0A0A9VT26 U5EZJ5 A0A195BVZ3 A0A034VDQ7 J9JQJ9 K7IR41 T1D614 A0A2H8TM44 T1DJG0 A0A2S2Q8F2 A0A2A3E0D4 A0A2S2NHL5 A0A2S2PX76 A0A0Q9WTY8 F5HMZ7 A0A1Y9IUX8 A0A1B0CIJ9 F5HMZ6 B4NCQ7 F5HMZ8
A0A212EI95 A0A146LQB0 A0A194QST9 A0A194PD08 A0A1Y1N919 A0A1W4WIK9 A0A1W4W6X5 A0A2P8Z2I9 D2A1V3 A0A1B6F8E6 N6U707 A0A1B6J3P2 A0A336LRQ8 A0A1W4WIC2 A0A182FRA0 V5GL93 A0A1B6KQA7 U4UN98 A0A1J1ID49 A0A1W4WHA8 A0A1A9W8U5 A0A1B6C8U4 A0A336K4G2 A0A1I8NPB4 A0A0A1WP88 A0A034V8T2 W8BKM4 A0A1I8NPB8 A0A0K8UWN4 A0A2M4CK37 A0A0K8W195 A0A2M3ZFF0 A0A1Q3EU33 A0A2M4BBK7 A0A0C9QZM2 A0A2M4A1I7 A0A3L8DEM8 A0A2M4B975 A0A0A1XLU7 A0A2M4CS68 A0A2M4CRQ0 A0A2M4CRW5 A0A2M4A0T0 A0A0C9Q5A0 A0A0A9VXE5 Q17FM4 A0A0K8TG95 A0A1S4G6S6 A0A0A9VT26 U5EZJ5 A0A195BVZ3 A0A034VDQ7 J9JQJ9 K7IR41 T1D614 A0A2H8TM44 T1DJG0 A0A2S2Q8F2 A0A2A3E0D4 A0A2S2NHL5 A0A2S2PX76 A0A0Q9WTY8 F5HMZ7 A0A1Y9IUX8 A0A1B0CIJ9 F5HMZ6 B4NCQ7 F5HMZ8
Ontologies
PANTHER
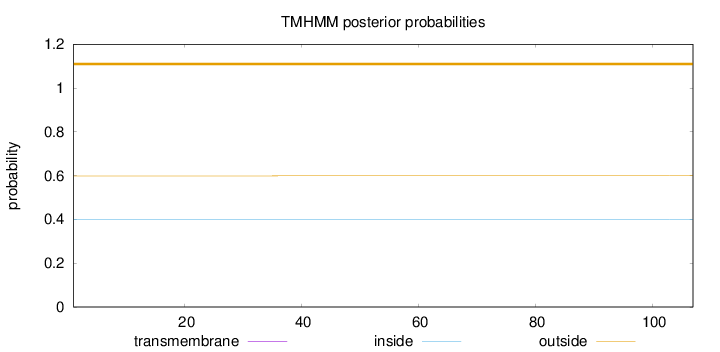
Topology
Length:
107
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00067
Exp number, first 60 AAs:
0.00067
Total prob of N-in:
0.40116
outside
1 - 107
Population Genetic Test Statistics
Pi
190.082815
Theta
187.097275
Tajima's D
-0.574266
CLR
0.972982
CSRT
0.229088545572721
Interpretation
Uncertain
