Pre Gene Modal
BGIBMGA008445
Annotation
PREDICTED:_indole-3-acetaldehyde_oxidase-like_isoform_X2_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.879
Sequence
CDS
ATGTCGCTAAATGCGTACATCAGATACGTGTGTGGTCTGACCGGGACTAAGGCGATGTGCCGTGAAGGAGGCTGCGGTGCTTGCATCGTCACAGTACGAGCCAAACGGTATCCCAGTGGAAAGACTGAAACATTTTCCGTAAATTCGTGTCTCGTGCTTGTATTTTCGTGTCACGGTTGGGATATAAGGACAATCGAAAGCACTGGTAATAGATTGGACGGGTACAGTGATATTCAGAAACTCACCGCTTGTTTACACGGAACCCAGTGTGGGTATTGCACACCCGGGTGGATTATGCACTTGAATAGGTGA
Protein
MSLNAYIRYVCGLTGTKAMCREGGCGACIVTVRAKRYPSGKTETFSVNSCLVLVFSCHGWDIRTIESTGNRLDGYSDIQKLTACLHGTQCGYCTPGWIMHLNR
Summary
Cofactor
FAD
Mo-molybdopterin
[2Fe-2S] cluster
Mo-molybdopterin
[2Fe-2S] cluster
Similarity
Belongs to the UDP-glycosyltransferase family.
Uniprot
A0A1U9X1S0
H9JFZ8
A0A2H1WDE7
A0A0H4TY60
A0A0F7QIE0
A0A076FRM9
+ More
A0A2H1WKN9 A0A2A4KA50 S5FPI8 A0A194PCA1 A0A1L8D6Y2 A0A212F2C6 A0A2H1VH28 A0A2H1V7P8 A0A076FX80 A0A1L8D6P5 A0A2A4JXM9 A0A2A4JX39 A0A2A4JYR8 H9JFZ3 A8TUC0 A0A0F7QJV6 A0A3S2NMY0 A0A0F7QEA1 A0A1X9ZM23 A0A076FRA1 A0A2A4JY06 A0A1X9ZLZ6 A0A194PDZ5 Q4VGM3 A0A194QM53 A0A194QMP0 A0A1L8D6Q4 H9JFZ2 A8TUB4 A0A0F7QEA3 A0A2H1VSX5 A0A212FKB8 A0A2A4IZZ7 A0A212F791 A0A0H4TD12 A0A067QPP3 A0A0A9YA60 A0A146L5G6 A0A0A9YHM8 A0A146MB53 A0A1Y9H2F5 A0A1X9ZM11 A0A0N1I7K2 A0A226F3L9 A0A1Y9HAI9 A0A336MA50 A0A1B6DXB5 A0A023F0C3 A0A3S2P6B3 U5EY22 A0A2R7WMZ6 A0A182MD19 A0A2R7WVC0 A0A0K8S633 A0A2H1VA49 T1HWZ8 A0A0K8S7B1 A0A0K1YWY0 A0A2J7PXR6 A0A1Y9H2G4 A0A1B0GL14 A0A2H1W7E2 A0A182RDV8 A0A182Q3A3 W5JEG5 J9HG89 A0A182SKH7 A0A0F7QIE4 A0A182YKY1 A0A1Y9HF24 A0A1D2NKQ2 T1I1X5 A0A194PIN2 A0A182R8Q6 A0A182JH14 T1P8E3 A0A182K1U4 A0A0A1XAA8 A0A1D2NIB3 A0A1D2NL30 A0A195E3I3 A0A1B6E3D0 A0A1I8M940 A0A182WLR0 A0A182V3I2 A0A182IIM6 A0A1B0CH66 A7UU59 A7UU58 A0A182LLJ7 A0A2M4CMF8 A0A1S4G5Y5 W5JB46 A0A2H1V6B6
A0A2H1WKN9 A0A2A4KA50 S5FPI8 A0A194PCA1 A0A1L8D6Y2 A0A212F2C6 A0A2H1VH28 A0A2H1V7P8 A0A076FX80 A0A1L8D6P5 A0A2A4JXM9 A0A2A4JX39 A0A2A4JYR8 H9JFZ3 A8TUC0 A0A0F7QJV6 A0A3S2NMY0 A0A0F7QEA1 A0A1X9ZM23 A0A076FRA1 A0A2A4JY06 A0A1X9ZLZ6 A0A194PDZ5 Q4VGM3 A0A194QM53 A0A194QMP0 A0A1L8D6Q4 H9JFZ2 A8TUB4 A0A0F7QEA3 A0A2H1VSX5 A0A212FKB8 A0A2A4IZZ7 A0A212F791 A0A0H4TD12 A0A067QPP3 A0A0A9YA60 A0A146L5G6 A0A0A9YHM8 A0A146MB53 A0A1Y9H2F5 A0A1X9ZM11 A0A0N1I7K2 A0A226F3L9 A0A1Y9HAI9 A0A336MA50 A0A1B6DXB5 A0A023F0C3 A0A3S2P6B3 U5EY22 A0A2R7WMZ6 A0A182MD19 A0A2R7WVC0 A0A0K8S633 A0A2H1VA49 T1HWZ8 A0A0K8S7B1 A0A0K1YWY0 A0A2J7PXR6 A0A1Y9H2G4 A0A1B0GL14 A0A2H1W7E2 A0A182RDV8 A0A182Q3A3 W5JEG5 J9HG89 A0A182SKH7 A0A0F7QIE4 A0A182YKY1 A0A1Y9HF24 A0A1D2NKQ2 T1I1X5 A0A194PIN2 A0A182R8Q6 A0A182JH14 T1P8E3 A0A182K1U4 A0A0A1XAA8 A0A1D2NIB3 A0A1D2NL30 A0A195E3I3 A0A1B6E3D0 A0A1I8M940 A0A182WLR0 A0A182V3I2 A0A182IIM6 A0A1B0CH66 A7UU59 A7UU58 A0A182LLJ7 A0A2M4CMF8 A0A1S4G5Y5 W5JB46 A0A2H1V6B6
Pubmed
EMBL
KY643683
AQY62686.1
BABH01001678
BABH01001679
ODYU01007852
SOQ50986.1
+ More
KP899547 AKQ06146.1 LC017754 BAR64768.1 KF960796 AII21998.1 ODYU01009314 SOQ53643.1 NWSH01000022 PCG80663.1 KC952900 AGQ43599.1 KQ459606 KPI90901.1 GEYN01000002 JAV02127.1 AGBW02010767 OWR47879.1 ODYU01002537 SOQ40155.1 ODYU01001109 SOQ36868.1 KF960794 AII21996.1 GEYN01000003 JAV02126.1 NWSH01000413 PCG76559.1 PCG76561.1 PCG76560.1 BABH01001661 EU073424 ABW71272.1 LC017753 BAR64767.1 RSAL01000229 RVE43893.1 LC017752 BAR64766.1 KY595779 ARS46833.1 KF960795 AII21997.1 PCG76558.1 KY595778 ARS46832.1 KPI90909.1 AY947538 AAY22446.1 KQ461196 KPJ06623.1 KPJ06614.1 GEYN01000001 JAV02128.1 BABH01001657 BABH01001658 BABH01001659 BABH01001660 EU073423 ABW71271.1 LC017757 BAR64771.1 ODYU01004253 SOQ43930.1 AGBW02008082 OWR54176.1 NWSH01004179 PCG65395.1 AGBW02009901 OWR49607.1 KP899546 AKQ06145.1 KK853740 KDQ87993.1 GBHO01014555 GBHO01014552 JAG29049.1 JAG29052.1 GDHC01015520 JAQ03109.1 GBHO01014557 GBHO01014554 GBRD01002756 JAG29047.1 JAG29050.1 JAG63065.1 GDHC01013650 GDHC01001960 JAQ04979.1 JAQ16669.1 KY595781 ARS46835.1 KQ460556 KPJ13979.1 LNIX01000001 OXA64068.1 AXCN02000553 AXCN02000554 UFQS01000745 UFQT01000745 SSX06553.1 SSX26900.1 GEDC01006998 JAS30300.1 GBBI01004413 JAC14299.1 RSAL01000295 RVE42846.1 GANO01002236 JAB57635.1 KK855082 PTY20711.1 AXCM01011484 KK855424 PTY22515.1 GBRD01017213 JAG48614.1 ODYU01001472 SOQ37709.1 ACPB03000354 ACPB03000355 GBRD01017212 JAG48615.1 KP859407 AKZ17717.1 NEVH01020852 PNF21139.1 AJWK01034051 ODYU01006502 SOQ48394.1 ADMH02001746 ETN61219.1 CH477659 EJY57867.1 LC017759 BAR64773.1 LJIJ01000015 ODN05821.1 ACPB03005165 KPI90950.1 KA644981 AFP59610.1 GBXI01006684 JAD07608.1 LJIJ01000032 ODN04991.1 ODN05815.1 KQ979701 KYN19698.1 GEDC01004862 JAS32436.1 APCN01000032 AJWK01012074 AJWK01012075 AAAB01008960 EDO63837.1 EDO63836.1 GGFL01002295 MBW66473.1 ETN61221.1 ODYU01000882 SOQ36316.1
KP899547 AKQ06146.1 LC017754 BAR64768.1 KF960796 AII21998.1 ODYU01009314 SOQ53643.1 NWSH01000022 PCG80663.1 KC952900 AGQ43599.1 KQ459606 KPI90901.1 GEYN01000002 JAV02127.1 AGBW02010767 OWR47879.1 ODYU01002537 SOQ40155.1 ODYU01001109 SOQ36868.1 KF960794 AII21996.1 GEYN01000003 JAV02126.1 NWSH01000413 PCG76559.1 PCG76561.1 PCG76560.1 BABH01001661 EU073424 ABW71272.1 LC017753 BAR64767.1 RSAL01000229 RVE43893.1 LC017752 BAR64766.1 KY595779 ARS46833.1 KF960795 AII21997.1 PCG76558.1 KY595778 ARS46832.1 KPI90909.1 AY947538 AAY22446.1 KQ461196 KPJ06623.1 KPJ06614.1 GEYN01000001 JAV02128.1 BABH01001657 BABH01001658 BABH01001659 BABH01001660 EU073423 ABW71271.1 LC017757 BAR64771.1 ODYU01004253 SOQ43930.1 AGBW02008082 OWR54176.1 NWSH01004179 PCG65395.1 AGBW02009901 OWR49607.1 KP899546 AKQ06145.1 KK853740 KDQ87993.1 GBHO01014555 GBHO01014552 JAG29049.1 JAG29052.1 GDHC01015520 JAQ03109.1 GBHO01014557 GBHO01014554 GBRD01002756 JAG29047.1 JAG29050.1 JAG63065.1 GDHC01013650 GDHC01001960 JAQ04979.1 JAQ16669.1 KY595781 ARS46835.1 KQ460556 KPJ13979.1 LNIX01000001 OXA64068.1 AXCN02000553 AXCN02000554 UFQS01000745 UFQT01000745 SSX06553.1 SSX26900.1 GEDC01006998 JAS30300.1 GBBI01004413 JAC14299.1 RSAL01000295 RVE42846.1 GANO01002236 JAB57635.1 KK855082 PTY20711.1 AXCM01011484 KK855424 PTY22515.1 GBRD01017213 JAG48614.1 ODYU01001472 SOQ37709.1 ACPB03000354 ACPB03000355 GBRD01017212 JAG48615.1 KP859407 AKZ17717.1 NEVH01020852 PNF21139.1 AJWK01034051 ODYU01006502 SOQ48394.1 ADMH02001746 ETN61219.1 CH477659 EJY57867.1 LC017759 BAR64773.1 LJIJ01000015 ODN05821.1 ACPB03005165 KPI90950.1 KA644981 AFP59610.1 GBXI01006684 JAD07608.1 LJIJ01000032 ODN04991.1 ODN05815.1 KQ979701 KYN19698.1 GEDC01004862 JAS32436.1 APCN01000032 AJWK01012074 AJWK01012075 AAAB01008960 EDO63837.1 EDO63836.1 GGFL01002295 MBW66473.1 ETN61221.1 ODYU01000882 SOQ36316.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000283053
UP000053240
+ More
UP000027135 UP000075884 UP000198287 UP000075886 UP000075883 UP000015103 UP000235965 UP000092461 UP000075900 UP000000673 UP000008820 UP000075901 UP000076408 UP000094527 UP000075880 UP000075881 UP000078492 UP000095301 UP000075920 UP000075903 UP000075840 UP000007062 UP000075882
UP000027135 UP000075884 UP000198287 UP000075886 UP000075883 UP000015103 UP000235965 UP000092461 UP000075900 UP000000673 UP000008820 UP000075901 UP000076408 UP000094527 UP000075880 UP000075881 UP000078492 UP000095301 UP000075920 UP000075903 UP000075840 UP000007062 UP000075882
Pfam
Interpro
IPR002888
2Fe-2S-bd
+ More
IPR016208 Ald_Oxase/xanthine_DH
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR036318 FAD-bd_PCMH-like_sf
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR016169 FAD-bd_PCMH_sub2
IPR012675 Beta-grasp_dom_sf
IPR002159 CD36_fam
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR035595 UDP_glycos_trans_CS
IPR013806 Kringle-like
IPR038178 Kringle_sf
IPR000001 Kringle
IPR041775 Ror-like_CRD
IPR020067 Frizzled_dom
IPR036790 Frizzled_dom_sf
IPR002213 UDP_glucos_trans
IPR016208 Ald_Oxase/xanthine_DH
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR036318 FAD-bd_PCMH-like_sf
IPR036884 2Fe-2S-bd_dom_sf
IPR005107 CO_DH_flav_C
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR002346 Mopterin_DH_FAD-bd
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR016169 FAD-bd_PCMH_sub2
IPR012675 Beta-grasp_dom_sf
IPR002159 CD36_fam
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR035595 UDP_glycos_trans_CS
IPR013806 Kringle-like
IPR038178 Kringle_sf
IPR000001 Kringle
IPR041775 Ror-like_CRD
IPR020067 Frizzled_dom
IPR036790 Frizzled_dom_sf
IPR002213 UDP_glucos_trans
SUPFAM
Gene 3D
ProteinModelPortal
A0A1U9X1S0
H9JFZ8
A0A2H1WDE7
A0A0H4TY60
A0A0F7QIE0
A0A076FRM9
+ More
A0A2H1WKN9 A0A2A4KA50 S5FPI8 A0A194PCA1 A0A1L8D6Y2 A0A212F2C6 A0A2H1VH28 A0A2H1V7P8 A0A076FX80 A0A1L8D6P5 A0A2A4JXM9 A0A2A4JX39 A0A2A4JYR8 H9JFZ3 A8TUC0 A0A0F7QJV6 A0A3S2NMY0 A0A0F7QEA1 A0A1X9ZM23 A0A076FRA1 A0A2A4JY06 A0A1X9ZLZ6 A0A194PDZ5 Q4VGM3 A0A194QM53 A0A194QMP0 A0A1L8D6Q4 H9JFZ2 A8TUB4 A0A0F7QEA3 A0A2H1VSX5 A0A212FKB8 A0A2A4IZZ7 A0A212F791 A0A0H4TD12 A0A067QPP3 A0A0A9YA60 A0A146L5G6 A0A0A9YHM8 A0A146MB53 A0A1Y9H2F5 A0A1X9ZM11 A0A0N1I7K2 A0A226F3L9 A0A1Y9HAI9 A0A336MA50 A0A1B6DXB5 A0A023F0C3 A0A3S2P6B3 U5EY22 A0A2R7WMZ6 A0A182MD19 A0A2R7WVC0 A0A0K8S633 A0A2H1VA49 T1HWZ8 A0A0K8S7B1 A0A0K1YWY0 A0A2J7PXR6 A0A1Y9H2G4 A0A1B0GL14 A0A2H1W7E2 A0A182RDV8 A0A182Q3A3 W5JEG5 J9HG89 A0A182SKH7 A0A0F7QIE4 A0A182YKY1 A0A1Y9HF24 A0A1D2NKQ2 T1I1X5 A0A194PIN2 A0A182R8Q6 A0A182JH14 T1P8E3 A0A182K1U4 A0A0A1XAA8 A0A1D2NIB3 A0A1D2NL30 A0A195E3I3 A0A1B6E3D0 A0A1I8M940 A0A182WLR0 A0A182V3I2 A0A182IIM6 A0A1B0CH66 A7UU59 A7UU58 A0A182LLJ7 A0A2M4CMF8 A0A1S4G5Y5 W5JB46 A0A2H1V6B6
A0A2H1WKN9 A0A2A4KA50 S5FPI8 A0A194PCA1 A0A1L8D6Y2 A0A212F2C6 A0A2H1VH28 A0A2H1V7P8 A0A076FX80 A0A1L8D6P5 A0A2A4JXM9 A0A2A4JX39 A0A2A4JYR8 H9JFZ3 A8TUC0 A0A0F7QJV6 A0A3S2NMY0 A0A0F7QEA1 A0A1X9ZM23 A0A076FRA1 A0A2A4JY06 A0A1X9ZLZ6 A0A194PDZ5 Q4VGM3 A0A194QM53 A0A194QMP0 A0A1L8D6Q4 H9JFZ2 A8TUB4 A0A0F7QEA3 A0A2H1VSX5 A0A212FKB8 A0A2A4IZZ7 A0A212F791 A0A0H4TD12 A0A067QPP3 A0A0A9YA60 A0A146L5G6 A0A0A9YHM8 A0A146MB53 A0A1Y9H2F5 A0A1X9ZM11 A0A0N1I7K2 A0A226F3L9 A0A1Y9HAI9 A0A336MA50 A0A1B6DXB5 A0A023F0C3 A0A3S2P6B3 U5EY22 A0A2R7WMZ6 A0A182MD19 A0A2R7WVC0 A0A0K8S633 A0A2H1VA49 T1HWZ8 A0A0K8S7B1 A0A0K1YWY0 A0A2J7PXR6 A0A1Y9H2G4 A0A1B0GL14 A0A2H1W7E2 A0A182RDV8 A0A182Q3A3 W5JEG5 J9HG89 A0A182SKH7 A0A0F7QIE4 A0A182YKY1 A0A1Y9HF24 A0A1D2NKQ2 T1I1X5 A0A194PIN2 A0A182R8Q6 A0A182JH14 T1P8E3 A0A182K1U4 A0A0A1XAA8 A0A1D2NIB3 A0A1D2NL30 A0A195E3I3 A0A1B6E3D0 A0A1I8M940 A0A182WLR0 A0A182V3I2 A0A182IIM6 A0A1B0CH66 A7UU59 A7UU58 A0A182LLJ7 A0A2M4CMF8 A0A1S4G5Y5 W5JB46 A0A2H1V6B6
PDB
1JRP
E-value=2.51494e-12,
Score=167
Ontologies
GO
Topology
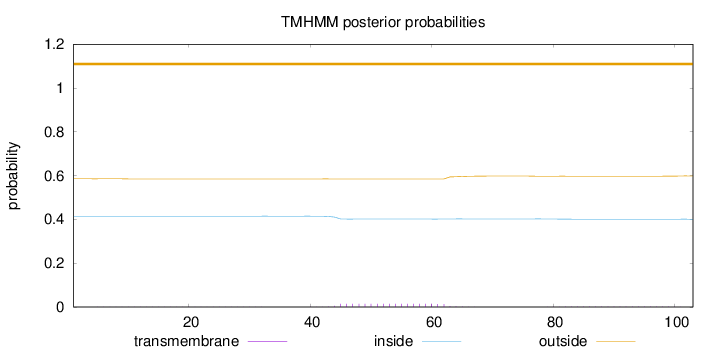
Length:
103
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31406
Exp number, first 60 AAs:
0.24388
Total prob of N-in:
0.41454
outside
1 - 103
Population Genetic Test Statistics
Pi
57.589766
Theta
147.052536
Tajima's D
-1.144841
CLR
1690.196852
CSRT
0.113294335283236
Interpretation
Uncertain
