Gene
KWMTBOMO10813
Pre Gene Modal
BGIBMGA008381
Annotation
UDP-glycosyltransferase_UGT50A1_precursor_[Bombyx_mori]
Full name
UDP-glucuronosyltransferase
Location in the cell
PlasmaMembrane Reliability : 2.944
Sequence
CDS
ATGGGAGGGACCAAGTCACATAAGATACCATTCTGGTCGCTTGCTGGAGGACTTACCCGAAGAGGACACAATATTACTTTCATAAGTGCGTTTCCACCTGATTTCCACATCGCTGGCTTAGAAGAAATAGCTCCAGAAGGTCTGGTATCATACGTTAAAAGCTACATGTCCTGGGACCTTGTTGGTGCTAGAATGAGAGGAGAGGAGCCTTTGCCAGCTTTTGATATACTTCGCTATGGATATGAGGCATGTGACGCACTACTCAAAGACTACGAGATGCGGTCTTTCCTGCAGTCAGGAAGAACTTACGATCTCATCATCATCGACGGAACGTATCCTGAGTGCGCGCTGGGGATCACATACAAAATGAAAGTGCCTTTCATGTACATTAACACGGTCGGCTTCTACTCGATGCCTTTGAGTAATGCAGGAAACCCAGCGCCATACTCCGTCACACCTTTCTTTGGAAGAGCCTTCACGGATAACATGGGAATCATTGAAAGAGCAATGAACTCCGCGTGGCAAATCGGTGCGATGGCGCTCCACGGTGTGAGCATGACCATACTTCAAGGGGTCTTGAGAAGACATTTCGGTAGCCAGATGCCACACGTGTACGATATGTCCAAGAACGTTAGCTTTATCCTCCAGAACGCGCATTATACTGTGTCATATCCGAGGCCTTATTTGCCGAATGTGGCTGAGATTGCCTGTATACATTGCATCGAACCGAAGAGATTGGATCCAGAAATAGAAGAATGGATTTCCGGTGCAGGCGACACCGGTTTTGTGTATGTCTCCATGGGATCTTCCGTGAAGACATCGAAAATGCCTTTAACAGCTCATCGGATGCTAATTAATGCTCTAGGAAGACTACCACAAAGGGTTCTATGGAAACAAGACGCGGTACAAAATATGACCGACATACCATCGAATGTGAAGCTATTGAAATGGTCACCACAGCAAGATTTACTTGGCCATCCAAAGATAAAGGCCTTCATCACCCACGGCGGTCTATTGAGCATGTTCGAAACGGTCTACCACGGCGTACCCATCGTCACCATACCGGTCTTCTGCGACCACGATGCCAACGCCGCGAAAGCAGAAGTCGACGGCTACGCTAAAAAACTAGAGTTCCAATATTTGACTTCTGACAAACTGCACGAAGCCATCCAAGAGGTTATAAATAATCCAAAATATAGGAGGGAAGTTAAATATCGACAGAATTTGCTTCGAGACCAGAAGGAGAGCCCTTTGGATAGGGCCGTGTACTGGACGGAATACGTCATAAGGCATAAAGGGGCCTATCATCTTCAATCTCCCGCAAAGGACCTCACCTTCATCCAATATTACTTGTTAGATGTGGCAATGCTATTTGTGATATCCGCTTTAGCCTTCTACGCTCTTATTTCCTTCGCGATAAGATCCAGTTTCCAAAGGCTCGCGGTTTTCATACAGAATCGACAAATGAAAATGTTATTCGATAATTCGACGGGACTGATTGGCAATTCTTTGATGGAGCAGAAAAAAAAATTGTGA
Protein
MGGTKSHKIPFWSLAGGLTRRGHNITFISAFPPDFHIAGLEEIAPEGLVSYVKSYMSWDLVGARMRGEEPLPAFDILRYGYEACDALLKDYEMRSFLQSGRTYDLIIIDGTYPECALGITYKMKVPFMYINTVGFYSMPLSNAGNPAPYSVTPFFGRAFTDNMGIIERAMNSAWQIGAMALHGVSMTILQGVLRRHFGSQMPHVYDMSKNVSFILQNAHYTVSYPRPYLPNVAEIACIHCIEPKRLDPEIEEWISGAGDTGFVYVSMGSSVKTSKMPLTAHRMLINALGRLPQRVLWKQDAVQNMTDIPSNVKLLKWSPQQDLLGHPKIKAFITHGGLLSMFETVYHGVPIVTIPVFCDHDANAAKAEVDGYAKKLEFQYLTSDKLHEAIQEVINNPKYRREVKYRQNLLRDQKESPLDRAVYWTEYVIRHKGAYHLQSPAKDLTFIQYYLLDVAMLFVISALAFYALISFAIRSSFQRLAVFIQNRQMKMLFDNSTGLIGNSLMEQKKKL
Summary
Catalytic Activity
glucuronate acceptor + UDP-alpha-D-glucuronate = acceptor beta-D-glucuronoside + H(+) + UDP
Similarity
Belongs to the UDP-glycosyltransferase family.
Feature
chain UDP-glucuronosyltransferase
Uniprot
G9LPW5
A0A194PD24
A0A194QSV5
G9LPS1
A0A2A4JX23
A0A2H1VJ78
+ More
A0A191T1Q2 A0A286MXP8 A0A3S2M2D3 A0A2H4WB76 A0A1L8D6G0 A0A212FKA0 A0A0L7LJ13 A0A1W4WHH1 A0A139WJ83 A0A1Y1M6B8 A0A1J1HU02 W8B991 A0A1I8NYU4 A0A2J7RLS9 A0A1A9WYI5 A0A182F8X6 B3N3K9 A0A0Q5WLQ3 A0A0Q5WPU1 A0A1B0GDH5 A0A1I8N3Z1 B4IWX8 A0A0L0BLI6 A0A1A9VY03 A0A182JEF6 A0A1A9YC12 A0A3B0JKS3 A0A084WQ49 A0A067RD76 A0A182VXB5 B4LGF5 A0A034W1U5 A0A182R9Z0 A0A0J9R602 A0A182G430 A0A182MMB2 B4KY17 A0A182NFI1 B4ISR1 A0A0K8UR33 A0A182USC7 A0A0A1WX88 A0A182HT17 A0A182Q6M1 B3MBR0 A0A182LF56 Q8SYL7 Q7QC46 B4MJ09 U4TVP7 A0A1A9ZX60 B5E0W3 A0A0R3NQS7 B4GBJ5 A0A0M4EEQ1 A0A182XWY5 Q17A89 A0A1B0GLB3 E0W3E2 A0A2P8YG55 Q7K142 B4QCM6 A0A182KF22 A0A182TZY8 A0A1I8N9Y4 A0A0L0CJN9 A0A1B6DDM3 A0A182TAG8 A0A182P752 A0A1B6MVC3 A0A1B6KCX6 A0A1W4W9X6 A0A1I8NVP3 B4KAR8 A0A1B6KXZ3 A0A0L7QP17 A0A154PNR0 A0A088AP08 A0A0J7NUR1 E9IDZ1 A0A195EU22 E2ATL2 A0A195DQA6 E2BR99 A0A2A3ES13 A0A151IIJ7 A0A0C9R7K0 A0A158NGI7 A0A195BXX1 A0A026W043
A0A191T1Q2 A0A286MXP8 A0A3S2M2D3 A0A2H4WB76 A0A1L8D6G0 A0A212FKA0 A0A0L7LJ13 A0A1W4WHH1 A0A139WJ83 A0A1Y1M6B8 A0A1J1HU02 W8B991 A0A1I8NYU4 A0A2J7RLS9 A0A1A9WYI5 A0A182F8X6 B3N3K9 A0A0Q5WLQ3 A0A0Q5WPU1 A0A1B0GDH5 A0A1I8N3Z1 B4IWX8 A0A0L0BLI6 A0A1A9VY03 A0A182JEF6 A0A1A9YC12 A0A3B0JKS3 A0A084WQ49 A0A067RD76 A0A182VXB5 B4LGF5 A0A034W1U5 A0A182R9Z0 A0A0J9R602 A0A182G430 A0A182MMB2 B4KY17 A0A182NFI1 B4ISR1 A0A0K8UR33 A0A182USC7 A0A0A1WX88 A0A182HT17 A0A182Q6M1 B3MBR0 A0A182LF56 Q8SYL7 Q7QC46 B4MJ09 U4TVP7 A0A1A9ZX60 B5E0W3 A0A0R3NQS7 B4GBJ5 A0A0M4EEQ1 A0A182XWY5 Q17A89 A0A1B0GLB3 E0W3E2 A0A2P8YG55 Q7K142 B4QCM6 A0A182KF22 A0A182TZY8 A0A1I8N9Y4 A0A0L0CJN9 A0A1B6DDM3 A0A182TAG8 A0A182P752 A0A1B6MVC3 A0A1B6KCX6 A0A1W4W9X6 A0A1I8NVP3 B4KAR8 A0A1B6KXZ3 A0A0L7QP17 A0A154PNR0 A0A088AP08 A0A0J7NUR1 E9IDZ1 A0A195EU22 E2ATL2 A0A195DQA6 E2BR99 A0A2A3ES13 A0A151IIJ7 A0A0C9R7K0 A0A158NGI7 A0A195BXX1 A0A026W043
EC Number
2.4.1.17
Pubmed
22155036
26354079
19954531
25299618
29027758
22118469
+ More
26227816 18362917 19820115 28004739 24495485 17994087 25315136 26108605 24438588 24845553 18057021 25348373 22936249 26483478 17550304 25830018 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 23537049 15632085 25244985 17510324 20566863 29403074 21282665 20798317 21347285 24508170 30249741
26227816 18362917 19820115 28004739 24495485 17994087 25315136 26108605 24438588 24845553 18057021 25348373 22936249 26483478 17550304 25830018 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12364791 23537049 15632085 25244985 17510324 20566863 29403074 21282665 20798317 21347285 24508170 30249741
EMBL
JQ070272
AEW43188.1
KQ459606
KPI90908.1
KQ461196
KPJ06621.1
+ More
JQ070228 AEW43144.1 NWSH01000413 PCG76557.1 ODYU01002865 SOQ40878.1 KU680311 ANI22017.1 MF684366 ASX94000.1 RSAL01000065 RVE49397.1 KY202944 AUC64288.1 GEYN01000083 JAV02046.1 AGBW02008082 OWR54174.1 JTDY01000994 KOB75166.1 KQ971338 KYB28033.1 GEZM01039714 GEZM01039713 JAV81274.1 CVRI01000021 CRK91551.1 GAMC01011354 GAMC01011353 JAB95202.1 NEVH01002683 PNF41797.1 CH954177 EDV59891.2 KQS71049.1 KQS71050.1 CCAG010011889 CH916366 EDV97379.1 JRES01001695 KNC20906.1 OUUW01000001 SPP75960.1 ATLV01025241 ATLV01025242 KE525377 KFB52343.1 KK852543 KDR21702.1 CH940647 EDW70484.1 KRF84936.1 GAKP01010650 JAC48302.1 CM002911 KMY91627.1 JXUM01041907 JXUM01041908 JXUM01041909 JXUM01041910 KQ561303 KXJ79059.1 AXCM01007871 CH933809 EDW18719.1 CH891581 EDW99493.1 GDHF01023160 JAI29154.1 GBXI01011056 JAD03236.1 APCN01000397 AXCN02000212 CH902619 EDV38206.1 AY071467 AE013599 AAL49089.1 AAM68364.3 AAAB01008859 EAA08100.5 CH963719 EDW72098.2 KB631735 ERL85659.1 CM000071 EDY68673.2 KRT01437.1 CH479181 EDW31290.1 CP012525 ALC44296.1 CH477338 EAT43164.1 AJWK01034711 AJWK01034712 AJWK01034713 DS235882 EEB20148.1 PYGN01000621 PSN43242.1 AY069392 AAL39537.1 AAM68363.2 CM000362 EDX05815.1 JRES01000310 KNC32417.1 GEDC01013505 JAS23793.1 GEBQ01000110 JAT39867.1 GEBQ01030668 JAT09309.1 CH933806 EDW16805.1 GEBQ01023706 JAT16271.1 KQ414840 KOC60367.1 KQ435007 KZC13493.1 LBMM01001540 KMQ96120.1 GL762556 EFZ21158.1 KQ981979 KYN31402.1 GL442629 EFN63208.1 KQ980612 KYN15100.1 GL449936 EFN81788.1 KZ288193 PBC34006.1 KQ977474 KYN02509.1 GBYB01012309 GBYB01012311 GBYB01012329 JAG82076.1 JAG82078.1 JAG82096.1 ADTU01001607 KQ976398 KYM92791.1 KK107519 QOIP01000009 EZA49387.1 RLU18565.1
JQ070228 AEW43144.1 NWSH01000413 PCG76557.1 ODYU01002865 SOQ40878.1 KU680311 ANI22017.1 MF684366 ASX94000.1 RSAL01000065 RVE49397.1 KY202944 AUC64288.1 GEYN01000083 JAV02046.1 AGBW02008082 OWR54174.1 JTDY01000994 KOB75166.1 KQ971338 KYB28033.1 GEZM01039714 GEZM01039713 JAV81274.1 CVRI01000021 CRK91551.1 GAMC01011354 GAMC01011353 JAB95202.1 NEVH01002683 PNF41797.1 CH954177 EDV59891.2 KQS71049.1 KQS71050.1 CCAG010011889 CH916366 EDV97379.1 JRES01001695 KNC20906.1 OUUW01000001 SPP75960.1 ATLV01025241 ATLV01025242 KE525377 KFB52343.1 KK852543 KDR21702.1 CH940647 EDW70484.1 KRF84936.1 GAKP01010650 JAC48302.1 CM002911 KMY91627.1 JXUM01041907 JXUM01041908 JXUM01041909 JXUM01041910 KQ561303 KXJ79059.1 AXCM01007871 CH933809 EDW18719.1 CH891581 EDW99493.1 GDHF01023160 JAI29154.1 GBXI01011056 JAD03236.1 APCN01000397 AXCN02000212 CH902619 EDV38206.1 AY071467 AE013599 AAL49089.1 AAM68364.3 AAAB01008859 EAA08100.5 CH963719 EDW72098.2 KB631735 ERL85659.1 CM000071 EDY68673.2 KRT01437.1 CH479181 EDW31290.1 CP012525 ALC44296.1 CH477338 EAT43164.1 AJWK01034711 AJWK01034712 AJWK01034713 DS235882 EEB20148.1 PYGN01000621 PSN43242.1 AY069392 AAL39537.1 AAM68363.2 CM000362 EDX05815.1 JRES01000310 KNC32417.1 GEDC01013505 JAS23793.1 GEBQ01000110 JAT39867.1 GEBQ01030668 JAT09309.1 CH933806 EDW16805.1 GEBQ01023706 JAT16271.1 KQ414840 KOC60367.1 KQ435007 KZC13493.1 LBMM01001540 KMQ96120.1 GL762556 EFZ21158.1 KQ981979 KYN31402.1 GL442629 EFN63208.1 KQ980612 KYN15100.1 GL449936 EFN81788.1 KZ288193 PBC34006.1 KQ977474 KYN02509.1 GBYB01012309 GBYB01012311 GBYB01012329 JAG82076.1 JAG82078.1 JAG82096.1 ADTU01001607 KQ976398 KYM92791.1 KK107519 QOIP01000009 EZA49387.1 RLU18565.1
Proteomes
UP000053268
UP000053240
UP000218220
UP000283053
UP000007151
UP000037510
+ More
UP000192223 UP000007266 UP000183832 UP000095300 UP000235965 UP000091820 UP000069272 UP000008711 UP000092444 UP000095301 UP000001070 UP000037069 UP000078200 UP000075880 UP000092443 UP000268350 UP000030765 UP000027135 UP000075920 UP000008792 UP000075900 UP000069940 UP000249989 UP000075883 UP000009192 UP000075884 UP000002282 UP000075903 UP000075840 UP000075886 UP000007801 UP000075882 UP000000803 UP000007062 UP000007798 UP000030742 UP000092445 UP000001819 UP000008744 UP000092553 UP000076408 UP000008820 UP000092461 UP000009046 UP000245037 UP000000304 UP000075881 UP000075902 UP000075901 UP000075885 UP000192221 UP000053825 UP000076502 UP000005203 UP000036403 UP000078541 UP000000311 UP000078492 UP000008237 UP000242457 UP000078542 UP000005205 UP000078540 UP000053097 UP000279307
UP000192223 UP000007266 UP000183832 UP000095300 UP000235965 UP000091820 UP000069272 UP000008711 UP000092444 UP000095301 UP000001070 UP000037069 UP000078200 UP000075880 UP000092443 UP000268350 UP000030765 UP000027135 UP000075920 UP000008792 UP000075900 UP000069940 UP000249989 UP000075883 UP000009192 UP000075884 UP000002282 UP000075903 UP000075840 UP000075886 UP000007801 UP000075882 UP000000803 UP000007062 UP000007798 UP000030742 UP000092445 UP000001819 UP000008744 UP000092553 UP000076408 UP000008820 UP000092461 UP000009046 UP000245037 UP000000304 UP000075881 UP000075902 UP000075901 UP000075885 UP000192221 UP000053825 UP000076502 UP000005203 UP000036403 UP000078541 UP000000311 UP000078492 UP000008237 UP000242457 UP000078542 UP000005205 UP000078540 UP000053097 UP000279307
Pfam
PF00201 UDPGT
ProteinModelPortal
G9LPW5
A0A194PD24
A0A194QSV5
G9LPS1
A0A2A4JX23
A0A2H1VJ78
+ More
A0A191T1Q2 A0A286MXP8 A0A3S2M2D3 A0A2H4WB76 A0A1L8D6G0 A0A212FKA0 A0A0L7LJ13 A0A1W4WHH1 A0A139WJ83 A0A1Y1M6B8 A0A1J1HU02 W8B991 A0A1I8NYU4 A0A2J7RLS9 A0A1A9WYI5 A0A182F8X6 B3N3K9 A0A0Q5WLQ3 A0A0Q5WPU1 A0A1B0GDH5 A0A1I8N3Z1 B4IWX8 A0A0L0BLI6 A0A1A9VY03 A0A182JEF6 A0A1A9YC12 A0A3B0JKS3 A0A084WQ49 A0A067RD76 A0A182VXB5 B4LGF5 A0A034W1U5 A0A182R9Z0 A0A0J9R602 A0A182G430 A0A182MMB2 B4KY17 A0A182NFI1 B4ISR1 A0A0K8UR33 A0A182USC7 A0A0A1WX88 A0A182HT17 A0A182Q6M1 B3MBR0 A0A182LF56 Q8SYL7 Q7QC46 B4MJ09 U4TVP7 A0A1A9ZX60 B5E0W3 A0A0R3NQS7 B4GBJ5 A0A0M4EEQ1 A0A182XWY5 Q17A89 A0A1B0GLB3 E0W3E2 A0A2P8YG55 Q7K142 B4QCM6 A0A182KF22 A0A182TZY8 A0A1I8N9Y4 A0A0L0CJN9 A0A1B6DDM3 A0A182TAG8 A0A182P752 A0A1B6MVC3 A0A1B6KCX6 A0A1W4W9X6 A0A1I8NVP3 B4KAR8 A0A1B6KXZ3 A0A0L7QP17 A0A154PNR0 A0A088AP08 A0A0J7NUR1 E9IDZ1 A0A195EU22 E2ATL2 A0A195DQA6 E2BR99 A0A2A3ES13 A0A151IIJ7 A0A0C9R7K0 A0A158NGI7 A0A195BXX1 A0A026W043
A0A191T1Q2 A0A286MXP8 A0A3S2M2D3 A0A2H4WB76 A0A1L8D6G0 A0A212FKA0 A0A0L7LJ13 A0A1W4WHH1 A0A139WJ83 A0A1Y1M6B8 A0A1J1HU02 W8B991 A0A1I8NYU4 A0A2J7RLS9 A0A1A9WYI5 A0A182F8X6 B3N3K9 A0A0Q5WLQ3 A0A0Q5WPU1 A0A1B0GDH5 A0A1I8N3Z1 B4IWX8 A0A0L0BLI6 A0A1A9VY03 A0A182JEF6 A0A1A9YC12 A0A3B0JKS3 A0A084WQ49 A0A067RD76 A0A182VXB5 B4LGF5 A0A034W1U5 A0A182R9Z0 A0A0J9R602 A0A182G430 A0A182MMB2 B4KY17 A0A182NFI1 B4ISR1 A0A0K8UR33 A0A182USC7 A0A0A1WX88 A0A182HT17 A0A182Q6M1 B3MBR0 A0A182LF56 Q8SYL7 Q7QC46 B4MJ09 U4TVP7 A0A1A9ZX60 B5E0W3 A0A0R3NQS7 B4GBJ5 A0A0M4EEQ1 A0A182XWY5 Q17A89 A0A1B0GLB3 E0W3E2 A0A2P8YG55 Q7K142 B4QCM6 A0A182KF22 A0A182TZY8 A0A1I8N9Y4 A0A0L0CJN9 A0A1B6DDM3 A0A182TAG8 A0A182P752 A0A1B6MVC3 A0A1B6KCX6 A0A1W4W9X6 A0A1I8NVP3 B4KAR8 A0A1B6KXZ3 A0A0L7QP17 A0A154PNR0 A0A088AP08 A0A0J7NUR1 E9IDZ1 A0A195EU22 E2ATL2 A0A195DQA6 E2BR99 A0A2A3ES13 A0A151IIJ7 A0A0C9R7K0 A0A158NGI7 A0A195BXX1 A0A026W043
PDB
2O6L
E-value=7.444e-31,
Score=335
Ontologies
GO
Topology
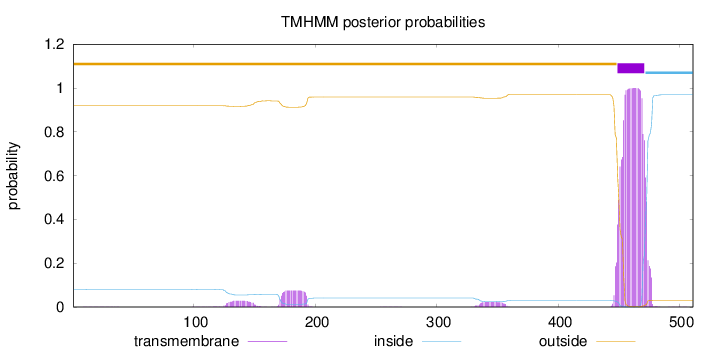
Subcellular location
Membrane
Length:
511
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.19704
Exp number, first 60 AAs:
0.00631
Total prob of N-in:
0.08087
outside
1 - 448
TMhelix
449 - 471
inside
472 - 511
Population Genetic Test Statistics
Pi
201.306617
Theta
151.112647
Tajima's D
1.809611
CLR
0.890563
CSRT
0.852907354632268
Interpretation
Uncertain
