Gene
KWMTBOMO10809
Pre Gene Modal
BGIBMGA008440
Annotation
aldehyde_oxidase_2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.531
Sequence
CDS
ATGATTGTAATAGATTGTGGTGATCATACTTGGTATAAATGTTATACTTTAAGTGATGTTTTTAGTGTCATAGAAAAGAGTACGGATTATAAATTGATTGCTGGAAATACAGGACAAGGAGTATATCACGTCCTGGATTATCCTAAGCTTGTCATTGATATTTCTAACGTAACAGATATAAGAGAATATGTCGTGGATGTTAATCTAACACTTGGTGCTGGAATGACTTTAACAGAAATGATGGAATTATTCCTTAAGCTTTCTGATGACAACGTAGACTTTAACTACTTGAAGGAGTTTCATGATCACATGGACTTGGTCGCTCATTTGCCAGTAAGGAATATCGGAACCATCGGTGGTAACTTGTACTTGAAACACTGCAACAAGGAATTCCAGTCAGATTTATTTTTGCTATTCGAGACTGTTGGAGCTATGATTACAATAGCCGAAAAAATGGACAAAATATCGACAATGTACCTTACAGATTTCCTCGAAACAGAAATGAAGGGAAAAATAATAATCAATGTGATGTTACCCCCCTTATCAAGTTCTACGAAGATCAAAACGTACAAGATAATGCCGAGATCACAAAACGCACACGCGATAGTCAACGCAGGATTTCTTTGGAAATTCAAACAAAATTCAAGTATACTGGAAAAAGCGACAATAGTATACGGTGGAATATCACCAAACTTTATTCATGCGTCAAAAACCGAGAGTATTTTATCAAACACAGATCCGTTCTCCGATGAAACTATACAGGAGGCATTGAAGACTTTATATGATGAAGTCAAGCCTGAAAACTCTCCAACAGAGCCATCTGCTTCTTATAGAAGAATGTTGGCTGTTTCTTTATATTACAAAGCATTAATAAGTCAATGTCCCGATGATCGAATCAACCCGAAGTATAAATCTGGCGGTAATGTTATTAAGCGGAATACGTCCAAAGGCACACAAACTTTCGAGACAGATGAAAATCTTTGGCCTTTAAATCAACCTGTGATGAAATTAGAAGCTTTAGCACAATGCAGCGGGGAAGCTACATTTGCCAATGATTTAAAAGGAGAATCAGACGAAGTATATGCTGCTTTTGTAACTGCTGACGTCAAACCTGGTAGTATTATTAGTGGCTTTGATACTACAGAAGCTTTTAAAATAGCAGGAGTTTCAGGATTTTATACCGCACAGGACATTCCCGGAAATAATTCATTTACACCAACCAACGCACCTTTGATTTTAGTGAATGAAGAAATATTGTGTTCTAAACAGGTCAAATACTATGGCGAACCAGCAGCGATAATAGTAGCGGATAGAGAAAAAACGGCCATTAAGGCTGCTAAACTAATATCGATAAAATATGAATCCATCAATAAAAATAAACCTGTACTAACAATTGATGACGCATTGAAAAGTCCAGATAAAGATACACGAATAACCAAAAACAATGTTATATATCCCGTCGAAGTTGGTCATGATGTGAAATGTATTATTTATGGGGAATTAAATATTGAAACACAGCATCACTTTTATATGGAGCCACAAACTTGCGTCGCAAAGAAAACCGAAGATGGCTTAGAAATATATTCTTCTACGCAATGGCTGGATTTGGCTAATATGGCTGTTGCACAATGTCTAAGCGTGCCCATAAATAGTGTGAACGTGATCATTAGAAGAGTTGGTGGTAGTTATGGAGGTAAAATTACAAGATCTAGTCAGATAGCATGTGGAGCAGCTCTTATTACGCATCTAACTGGAAAAACATGCAGGTTTATTCTGCCTTTGCAACAAAATATGGGAATTATTGGTAAACGGCTCCCAACAAAATGTAATTTCGAAGTGGGTGTTGACAATAACGGTGAAATACAATACCTTAAAAATATATTCTATCAAGACAATGGATGTGCACCGAATGAAACGATTAGTCCAGTGACTGCAGCCCATTTTGTTGGGAACTGCTATGATAGTCGAAGATGGTACGTCGAAGCTAATAGCGCAGCAACTGATAGCCCATCTAACACTTGGTGTAGAGCTCCTTCTTCAACAGAAGCAATAGCAATGTGTGAATACATAATGGAAAAGGTAGCATATCATCTGAATAAAGATCCTTTAGAAGTGAGACTAACTAATATGATGCAAGTCACAAATCCTATTCCACAATTAATTGACCAACTAAAAAGAGACTCGGACTACGACCAAAGAATAATTGATGTGCAAAATTACAATAAGCAGAATAGATGGAGAAAGCGAGCTTTGAAACTCTTGCCAATGACTTATGATGTCTTTTATTTCGGGTCATATAATTCCGTTGTATCAGTTTATCACGCAGATGGGTCAGTTGTGATAATACATGGTGGAGTTGAAATGGGTCAAGGTTTAAACACAAAAGTAGCTCAAGTATGTGCATACATTTTTGGTATTCCCCTCAACAAAATTAGCGTAAAACCTAGTACAAGCTTTACGAGTCCAAACGCAATGACCACAGGTGGTAGTATTGGTAGTGAATGTGTGTCTTTTGCAACAATGAAAGCCTGCCAGATAATAATGGATCGCCTCAAACCAATCAAAGAAGAATTGAATGATCCAAAATGGGAGGATATTATTAAAAAAGCTTTCAATAACGATATAGATTTGCAAGCGTCATATATGTATTCAAATAAAGATGGCTTAAAGCCATATGATGTGTATGGTGTAGTTGTGATGGAGGTTGAAGTTGATATTTTAACTGGAAACCATGATGTGCTGAGAGTTGATTTACTGGAAGATACTGGCAGAAGTATGAGTCCGGGAATTGACGTTGGACAGATAGAGGGTGCGTTCATCATGGGTCTCGGCTATTGGACATCGGAAAAAGTGATTTATGATCAGGAAACTGGAAAGCTCCTAACGGATCGTACTTGGACGTACAAGCCTCCTGGTCTTAAGGATATACCGGCAGATTTCCGAATATATTTCAGGAGGAATTCCAACAACCCTACCGGAGTACTACAATCTAAAGCAACTGGCGAACCGGCATTCTCTCTAGCTGCAGTAATAACTCATGCTATACGGGATGCAGTTAGGGCTGCTAGACTTGACGCTGCCTACGAAGACCAATGGATTGATATTCCTAATCCCTGCACGGTGGAAAACATATTTATGGCTGTCGGACACAGATTAGACCAGTTCGTGTTGAAGTGA
Protein
MIVIDCGDHTWYKCYTLSDVFSVIEKSTDYKLIAGNTGQGVYHVLDYPKLVIDISNVTDIREYVVDVNLTLGAGMTLTEMMELFLKLSDDNVDFNYLKEFHDHMDLVAHLPVRNIGTIGGNLYLKHCNKEFQSDLFLLFETVGAMITIAEKMDKISTMYLTDFLETEMKGKIIINVMLPPLSSSTKIKTYKIMPRSQNAHAIVNAGFLWKFKQNSSILEKATIVYGGISPNFIHASKTESILSNTDPFSDETIQEALKTLYDEVKPENSPTEPSASYRRMLAVSLYYKALISQCPDDRINPKYKSGGNVIKRNTSKGTQTFETDENLWPLNQPVMKLEALAQCSGEATFANDLKGESDEVYAAFVTADVKPGSIISGFDTTEAFKIAGVSGFYTAQDIPGNNSFTPTNAPLILVNEEILCSKQVKYYGEPAAIIVADREKTAIKAAKLISIKYESINKNKPVLTIDDALKSPDKDTRITKNNVIYPVEVGHDVKCIIYGELNIETQHHFYMEPQTCVAKKTEDGLEIYSSTQWLDLANMAVAQCLSVPINSVNVIIRRVGGSYGGKITRSSQIACGAALITHLTGKTCRFILPLQQNMGIIGKRLPTKCNFEVGVDNNGEIQYLKNIFYQDNGCAPNETISPVTAAHFVGNCYDSRRWYVEANSAATDSPSNTWCRAPSSTEAIAMCEYIMEKVAYHLNKDPLEVRLTNMMQVTNPIPQLIDQLKRDSDYDQRIIDVQNYNKQNRWRKRALKLLPMTYDVFYFGSYNSVVSVYHADGSVVIIHGGVEMGQGLNTKVAQVCAYIFGIPLNKISVKPSTSFTSPNAMTTGGSIGSECVSFATMKACQIIMDRLKPIKEELNDPKWEDIIKKAFNNDIDLQASYMYSNKDGLKPYDVYGVVVMEVEVDILTGNHDVLRVDLLEDTGRSMSPGIDVGQIEGAFIMGLGYWTSEKVIYDQETGKLLTDRTWTYKPPGLKDIPADFRIYFRRNSNNPTGVLQSKATGEPAFSLAAVITHAIRDAVRAARLDAAYEDQWIDIPNPCTVENIFMAVGHRLDQFVLK
Summary
Cofactor
FAD
Mo-molybdopterin
[2Fe-2S] cluster
Mo-molybdopterin
[2Fe-2S] cluster
Similarity
Belongs to the UDP-glycosyltransferase family.
Uniprot
A8TUC0
A0A0F7QJV6
A0A3S2NMY0
A0A076FRA1
A0A2A4JY06
A0A1X9ZM23
+ More
A0A076FX80 A0A2A4JXM9 A0A212FKB8 A0A2H1VH28 H9JFZ2 A0A2A4JX39 A8TUB4 A0A194QM53 A0A2H1V7P8 A0A0F7QEA1 A0A1X9ZLZ6 A0A2A4JYR8 A0A194PDZ5 A0A2H1VCW8 A0A2H1VSX5 A0A194PIN2 A0A0H4TD12 A0A0F7QEA3 H9JFX4 A0A0L7LLP8 B0X3W4 H9JFX1 A0A194QBY5 A0A0N0BES0 A0A0F7QIE0 A0A2A4JB30 A0A1J1J970 A0A1Q3FGZ2 A0A1Q3FG61 A0A1X9ZM11 A0A182SKH7 E2ANE4 A0A182QVB6 A0A194QMP0 Q16T45 B0X3W2 A0A0H4TY60 S5FPI8 A0A2M4BA48 A0A212F2C6 A0A2M4BAX5 A0A2M4BAZ2 A0A0L7R0P8 W5JB46 A0A2M3ZFL0 A0A2M4BAH9 A0A1Q3FSG4 A0A240PNF0 A0A1Y9J0F9 A0A2J7R4N3 A0A194PE80 A0A1B6E3D0 A0A2M4CMF8 A0A182LLJ7 A0A2M4B9W4 A0A182RDV8 A0A1U9X1S0 A0A182Q3A3 A0A182IIM6 B0X3X6 A0A182J4M1 W5JEG5 A0A2M4CMZ5 A0A084WK44 A0A194PIT3 A0A023F0C3 A0A0N0PF58 A0A182YKY1 A0A2A4ITG3 A0A154PJ92 A0A3S2PBH0 A0A2H1V2I4 A0A336MA50 A0A1Y9H2G4 A0A182VPD9 A0A182WLR0 A0A1S4FQ35 U5EY22 A0A182GN26 A0A195FSQ5 Q7Q5T1 A0A310SF38 F4WWU5 A0A182VKT2 B0XER7 Q16T47 A0A182II50 A0A194PCQ5 A0A1L8DXU8 A0A151X416 A0A0N0PD45 A0A1L8DXS0 A0A151I2B1
A0A076FX80 A0A2A4JXM9 A0A212FKB8 A0A2H1VH28 H9JFZ2 A0A2A4JX39 A8TUB4 A0A194QM53 A0A2H1V7P8 A0A0F7QEA1 A0A1X9ZLZ6 A0A2A4JYR8 A0A194PDZ5 A0A2H1VCW8 A0A2H1VSX5 A0A194PIN2 A0A0H4TD12 A0A0F7QEA3 H9JFX4 A0A0L7LLP8 B0X3W4 H9JFX1 A0A194QBY5 A0A0N0BES0 A0A0F7QIE0 A0A2A4JB30 A0A1J1J970 A0A1Q3FGZ2 A0A1Q3FG61 A0A1X9ZM11 A0A182SKH7 E2ANE4 A0A182QVB6 A0A194QMP0 Q16T45 B0X3W2 A0A0H4TY60 S5FPI8 A0A2M4BA48 A0A212F2C6 A0A2M4BAX5 A0A2M4BAZ2 A0A0L7R0P8 W5JB46 A0A2M3ZFL0 A0A2M4BAH9 A0A1Q3FSG4 A0A240PNF0 A0A1Y9J0F9 A0A2J7R4N3 A0A194PE80 A0A1B6E3D0 A0A2M4CMF8 A0A182LLJ7 A0A2M4B9W4 A0A182RDV8 A0A1U9X1S0 A0A182Q3A3 A0A182IIM6 B0X3X6 A0A182J4M1 W5JEG5 A0A2M4CMZ5 A0A084WK44 A0A194PIT3 A0A023F0C3 A0A0N0PF58 A0A182YKY1 A0A2A4ITG3 A0A154PJ92 A0A3S2PBH0 A0A2H1V2I4 A0A336MA50 A0A1Y9H2G4 A0A182VPD9 A0A182WLR0 A0A1S4FQ35 U5EY22 A0A182GN26 A0A195FSQ5 Q7Q5T1 A0A310SF38 F4WWU5 A0A182VKT2 B0XER7 Q16T47 A0A182II50 A0A194PCQ5 A0A1L8DXU8 A0A151X416 A0A0N0PD45 A0A1L8DXS0 A0A151I2B1
Pubmed
EMBL
EU073424
ABW71272.1
LC017753
BAR64767.1
RSAL01000229
RVE43893.1
+ More
KF960795 AII21997.1 NWSH01000413 PCG76558.1 KY595779 ARS46833.1 KF960794 AII21996.1 PCG76559.1 AGBW02008082 OWR54176.1 ODYU01002537 SOQ40155.1 BABH01001657 BABH01001658 BABH01001659 BABH01001660 PCG76561.1 EU073423 ABW71271.1 KQ461196 KPJ06623.1 ODYU01001109 SOQ36868.1 LC017752 BAR64766.1 KY595778 ARS46832.1 PCG76560.1 KQ459606 KPI90909.1 ODYU01001878 SOQ38698.1 ODYU01004253 SOQ43930.1 KPI90950.1 KP899546 AKQ06145.1 LC017757 BAR64771.1 BABH01001568 BABH01001569 BABH01001570 BABH01001571 BABH01001572 BABH01001573 JTDY01000607 KOB76483.1 DS232324 EDS40041.1 BABH01001558 KQ459193 KPJ03063.1 KQ435826 KOX72014.1 LC017754 BAR64768.1 NWSH01002143 PCG69059.1 CVRI01000075 CRL08602.1 GFDL01008214 JAV26831.1 GFDL01008505 JAV26540.1 KY595781 ARS46835.1 GL441193 EFN65044.1 AXCN02000553 KPJ06614.1 CH477659 EAT37664.1 EDS40039.1 KP899547 AKQ06146.1 KC952900 AGQ43599.1 GGFJ01000772 MBW49913.1 AGBW02010767 OWR47879.1 GGFJ01001064 MBW50205.1 GGFJ01001065 MBW50206.1 KQ414670 KOC64413.1 ADMH02001746 ETN61221.1 GGFM01006571 MBW27322.1 GGFJ01000687 MBW49828.1 GFDL01004496 JAV30549.1 NEVH01007404 PNF35793.1 KPI90999.1 GEDC01004862 JAS32436.1 GGFL01002295 MBW66473.1 GGFJ01000686 MBW49827.1 KY643683 AQY62686.1 AXCN02000554 APCN01000032 EDS40053.1 ETN61219.1 GGFL01002475 MBW66653.1 ATLV01024093 KE525349 KFB50588.1 KPI91000.1 GBBI01004413 JAC14299.1 KQ459674 KPJ20482.1 NWSH01007172 PCG63055.1 KQ434912 KZC11368.1 RSAL01000123 RVE46648.1 ODYU01000376 SOQ35055.1 UFQS01000745 UFQT01000745 SSX06553.1 SSX26900.1 GANO01002236 JAB57635.1 JXUM01014357 JXUM01014358 JXUM01014359 KQ560401 KXJ82608.1 KQ981281 KYN43332.1 AAAB01008960 EAA11584.4 KQ769812 OAD52839.1 GL888413 EGI61333.1 DS232859 EDS26176.1 EAT37662.1 APCN01000031 KPI90997.1 GFDF01002797 JAV11287.1 KQ982557 KYQ55014.1 KQ460326 KPJ15694.1 GFDF01002843 JAV11241.1 KQ976546 KYM80948.1
KF960795 AII21997.1 NWSH01000413 PCG76558.1 KY595779 ARS46833.1 KF960794 AII21996.1 PCG76559.1 AGBW02008082 OWR54176.1 ODYU01002537 SOQ40155.1 BABH01001657 BABH01001658 BABH01001659 BABH01001660 PCG76561.1 EU073423 ABW71271.1 KQ461196 KPJ06623.1 ODYU01001109 SOQ36868.1 LC017752 BAR64766.1 KY595778 ARS46832.1 PCG76560.1 KQ459606 KPI90909.1 ODYU01001878 SOQ38698.1 ODYU01004253 SOQ43930.1 KPI90950.1 KP899546 AKQ06145.1 LC017757 BAR64771.1 BABH01001568 BABH01001569 BABH01001570 BABH01001571 BABH01001572 BABH01001573 JTDY01000607 KOB76483.1 DS232324 EDS40041.1 BABH01001558 KQ459193 KPJ03063.1 KQ435826 KOX72014.1 LC017754 BAR64768.1 NWSH01002143 PCG69059.1 CVRI01000075 CRL08602.1 GFDL01008214 JAV26831.1 GFDL01008505 JAV26540.1 KY595781 ARS46835.1 GL441193 EFN65044.1 AXCN02000553 KPJ06614.1 CH477659 EAT37664.1 EDS40039.1 KP899547 AKQ06146.1 KC952900 AGQ43599.1 GGFJ01000772 MBW49913.1 AGBW02010767 OWR47879.1 GGFJ01001064 MBW50205.1 GGFJ01001065 MBW50206.1 KQ414670 KOC64413.1 ADMH02001746 ETN61221.1 GGFM01006571 MBW27322.1 GGFJ01000687 MBW49828.1 GFDL01004496 JAV30549.1 NEVH01007404 PNF35793.1 KPI90999.1 GEDC01004862 JAS32436.1 GGFL01002295 MBW66473.1 GGFJ01000686 MBW49827.1 KY643683 AQY62686.1 AXCN02000554 APCN01000032 EDS40053.1 ETN61219.1 GGFL01002475 MBW66653.1 ATLV01024093 KE525349 KFB50588.1 KPI91000.1 GBBI01004413 JAC14299.1 KQ459674 KPJ20482.1 NWSH01007172 PCG63055.1 KQ434912 KZC11368.1 RSAL01000123 RVE46648.1 ODYU01000376 SOQ35055.1 UFQS01000745 UFQT01000745 SSX06553.1 SSX26900.1 GANO01002236 JAB57635.1 JXUM01014357 JXUM01014358 JXUM01014359 KQ560401 KXJ82608.1 KQ981281 KYN43332.1 AAAB01008960 EAA11584.4 KQ769812 OAD52839.1 GL888413 EGI61333.1 DS232859 EDS26176.1 EAT37662.1 APCN01000031 KPI90997.1 GFDF01002797 JAV11287.1 KQ982557 KYQ55014.1 KQ460326 KPJ15694.1 GFDF01002843 JAV11241.1 KQ976546 KYM80948.1
Proteomes
UP000283053
UP000218220
UP000007151
UP000005204
UP000053240
UP000053268
+ More
UP000037510 UP000002320 UP000053105 UP000183832 UP000075901 UP000000311 UP000075886 UP000008820 UP000053825 UP000000673 UP000075885 UP000076407 UP000235965 UP000075882 UP000075900 UP000075840 UP000075880 UP000030765 UP000076408 UP000076502 UP000075884 UP000075903 UP000075920 UP000069940 UP000249989 UP000078541 UP000007062 UP000007755 UP000075809 UP000078540
UP000037510 UP000002320 UP000053105 UP000183832 UP000075901 UP000000311 UP000075886 UP000008820 UP000053825 UP000000673 UP000075885 UP000076407 UP000235965 UP000075882 UP000075900 UP000075840 UP000075880 UP000030765 UP000076408 UP000076502 UP000075884 UP000075903 UP000075920 UP000069940 UP000249989 UP000078541 UP000007062 UP000007755 UP000075809 UP000078540
Pfam
Interpro
IPR016208
Ald_Oxase/xanthine_DH
+ More
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR002888 2Fe-2S-bd
IPR036884 2Fe-2S-bd_dom_sf
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR005107 CO_DH_flav_C
IPR036318 FAD-bd_PCMH-like_sf
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR002346 Mopterin_DH_FAD-bd
IPR016169 FAD-bd_PCMH_sub2
IPR012675 Beta-grasp_dom_sf
IPR002159 CD36_fam
IPR006621 Nose-resist-to-fluoxetine_N
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR013809 ENTH
IPR008942 ENTH_VHS
IPR011417 ANTH_dom
IPR014712 Clathrin_AP_dom2
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR002888 2Fe-2S-bd
IPR036884 2Fe-2S-bd_dom_sf
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR005107 CO_DH_flav_C
IPR036318 FAD-bd_PCMH-like_sf
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR002346 Mopterin_DH_FAD-bd
IPR016169 FAD-bd_PCMH_sub2
IPR012675 Beta-grasp_dom_sf
IPR002159 CD36_fam
IPR006621 Nose-resist-to-fluoxetine_N
IPR002213 UDP_glucos_trans
IPR035595 UDP_glycos_trans_CS
IPR013809 ENTH
IPR008942 ENTH_VHS
IPR011417 ANTH_dom
IPR014712 Clathrin_AP_dom2
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A8TUC0
A0A0F7QJV6
A0A3S2NMY0
A0A076FRA1
A0A2A4JY06
A0A1X9ZM23
+ More
A0A076FX80 A0A2A4JXM9 A0A212FKB8 A0A2H1VH28 H9JFZ2 A0A2A4JX39 A8TUB4 A0A194QM53 A0A2H1V7P8 A0A0F7QEA1 A0A1X9ZLZ6 A0A2A4JYR8 A0A194PDZ5 A0A2H1VCW8 A0A2H1VSX5 A0A194PIN2 A0A0H4TD12 A0A0F7QEA3 H9JFX4 A0A0L7LLP8 B0X3W4 H9JFX1 A0A194QBY5 A0A0N0BES0 A0A0F7QIE0 A0A2A4JB30 A0A1J1J970 A0A1Q3FGZ2 A0A1Q3FG61 A0A1X9ZM11 A0A182SKH7 E2ANE4 A0A182QVB6 A0A194QMP0 Q16T45 B0X3W2 A0A0H4TY60 S5FPI8 A0A2M4BA48 A0A212F2C6 A0A2M4BAX5 A0A2M4BAZ2 A0A0L7R0P8 W5JB46 A0A2M3ZFL0 A0A2M4BAH9 A0A1Q3FSG4 A0A240PNF0 A0A1Y9J0F9 A0A2J7R4N3 A0A194PE80 A0A1B6E3D0 A0A2M4CMF8 A0A182LLJ7 A0A2M4B9W4 A0A182RDV8 A0A1U9X1S0 A0A182Q3A3 A0A182IIM6 B0X3X6 A0A182J4M1 W5JEG5 A0A2M4CMZ5 A0A084WK44 A0A194PIT3 A0A023F0C3 A0A0N0PF58 A0A182YKY1 A0A2A4ITG3 A0A154PJ92 A0A3S2PBH0 A0A2H1V2I4 A0A336MA50 A0A1Y9H2G4 A0A182VPD9 A0A182WLR0 A0A1S4FQ35 U5EY22 A0A182GN26 A0A195FSQ5 Q7Q5T1 A0A310SF38 F4WWU5 A0A182VKT2 B0XER7 Q16T47 A0A182II50 A0A194PCQ5 A0A1L8DXU8 A0A151X416 A0A0N0PD45 A0A1L8DXS0 A0A151I2B1
A0A076FX80 A0A2A4JXM9 A0A212FKB8 A0A2H1VH28 H9JFZ2 A0A2A4JX39 A8TUB4 A0A194QM53 A0A2H1V7P8 A0A0F7QEA1 A0A1X9ZLZ6 A0A2A4JYR8 A0A194PDZ5 A0A2H1VCW8 A0A2H1VSX5 A0A194PIN2 A0A0H4TD12 A0A0F7QEA3 H9JFX4 A0A0L7LLP8 B0X3W4 H9JFX1 A0A194QBY5 A0A0N0BES0 A0A0F7QIE0 A0A2A4JB30 A0A1J1J970 A0A1Q3FGZ2 A0A1Q3FG61 A0A1X9ZM11 A0A182SKH7 E2ANE4 A0A182QVB6 A0A194QMP0 Q16T45 B0X3W2 A0A0H4TY60 S5FPI8 A0A2M4BA48 A0A212F2C6 A0A2M4BAX5 A0A2M4BAZ2 A0A0L7R0P8 W5JB46 A0A2M3ZFL0 A0A2M4BAH9 A0A1Q3FSG4 A0A240PNF0 A0A1Y9J0F9 A0A2J7R4N3 A0A194PE80 A0A1B6E3D0 A0A2M4CMF8 A0A182LLJ7 A0A2M4B9W4 A0A182RDV8 A0A1U9X1S0 A0A182Q3A3 A0A182IIM6 B0X3X6 A0A182J4M1 W5JEG5 A0A2M4CMZ5 A0A084WK44 A0A194PIT3 A0A023F0C3 A0A0N0PF58 A0A182YKY1 A0A2A4ITG3 A0A154PJ92 A0A3S2PBH0 A0A2H1V2I4 A0A336MA50 A0A1Y9H2G4 A0A182VPD9 A0A182WLR0 A0A1S4FQ35 U5EY22 A0A182GN26 A0A195FSQ5 Q7Q5T1 A0A310SF38 F4WWU5 A0A182VKT2 B0XER7 Q16T47 A0A182II50 A0A194PCQ5 A0A1L8DXU8 A0A151X416 A0A0N0PD45 A0A1L8DXS0 A0A151I2B1
PDB
3UNI
E-value=2.06157e-102,
Score=955
Ontologies
PATHWAY
GO
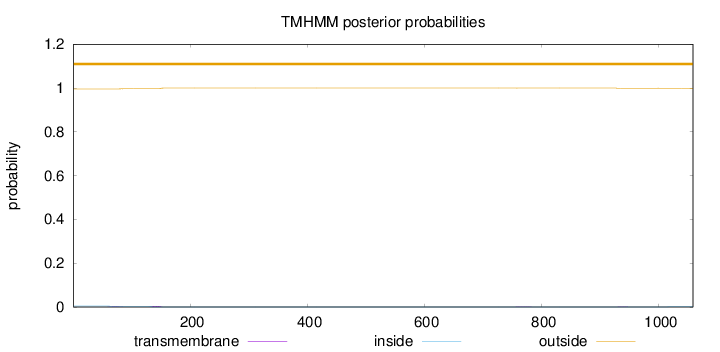
Topology
Length:
1058
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0949400000000001
Exp number, first 60 AAs:
0.00046
Total prob of N-in:
0.00428
outside
1 - 1058
Population Genetic Test Statistics
Pi
162.440639
Theta
171.421685
Tajima's D
-0.64475
CLR
27.63551
CSRT
0.206739663016849
Interpretation
Uncertain
