Gene
KWMTBOMO10799
Annotation
PREDICTED:_uncharacterized_protein_LOC101738890_[Bombyx_mori]
Full name
Retrovirus-related Pol polyprotein from transposon 17.6
Location in the cell
Nuclear Reliability : 2.36
Sequence
CDS
ATGCTCGACCAAAAAATCATACAACCTTCTCTATCTCCTTGGTCCTCGCCTATATGGATTGTCCCAAAAAAGACCGATGCCTCTGGTCAAAAGAAATGGCGGATTGTAATTGATTATAGGAAACTTAACGATATTACTATTGGTGAAACTTACCCCATACCTAACATTGTCGAAATTCTTGACCAATTAGGAAACTCAAAGTACTTCACAACATTAGATCTCGCTTCTGGCTTTCATCAGATAACAATGAATCAAAAAGACGCTTGCAAAACCGCGTTTAGTGTCCCTCAAGGACACTTTGAATTTACAAGGATGCCCTTTGGCCTAAAAAATGCCCCATCAACTTTTCAGCGCTTAATGAACAATGTGCTCACTGGCCTACAAGGTGAGCGCTGTTTTGTGTATTTAGACGATATTGTGTCTTATTCTCATGACCTTAAAACTCACATTGAAAACCTCGGTGCAATATTCGATAGACTACGTAAATTTAACCTAAAATTACAGCCAGACAAATGTGAATTTCTACGAAAAGAAGTCGGCTACCTAGGTCATATAATTAGCGAGGAAGGACTAAAACCAGACCCTAATAAAATAAAATCCGTTAAAGAGTTTCCAATCCCAAAATCCCCCAAGGACATTAAATCCTTTTTAGGACTAATATCATACTATAGACGCTTTATCCCAGAGTTCTCCAAACTATCAAAACCACTCACATCCCTCTTGAAAAAGGATGCATCTTTTCTTTGGACCAACGAACAACAATTAGCTTTCGAAACGCTCAAAGATAAACTCATTACCTCTCCTGTATTAATATACCCAGATTTTACTAAACCATTCAACCTTACTTGCGATGCATCGAACTATGCAATATCAGCTATCCTTTCGCAAGGTCCTATAGGTAAAGACCGCCCTGTAGCCTATGCATCCAGAACACTAAATAAATGTGAAATTAACTATAGCACAACCGAAAAGGAGTTGCTAGCTATAGTTTGGGGCTGCAATTCTTTTAGACCGTATTTATTTGGTCGTAAGTTCATCATAGTAACAGATCATCGTCCACTAAAATGGCTATTTAACCATACTGATCCAAGTAGCAAACTACAACGTTGGCGTCTCCAACTACAAGAATTTGAATACGAAATTATATATAGAAAAGGCAAATTAAATTCAGCAGCTGACGCTCTATCTCGTTATCCCGTTAATCCTGTCTATACCGAAAATCCTGAAAACTTAAATGAAAATGAACTGATTGATCTGGGTCCATTAGACATCAGTCCCTTTAATGACGAAGTCCCTGATTTTACGCCTCTAAGTCCAAGTCCTAACAATTTACCTGATACTCTGCTCCAACCAGAAATTGTACCCGAAATCACTGATCAAAATCCAATAATAGAAAATGAACCCCAACCTTCTAGTAGTCAAGCTCCTATTCTACAACAACCTAATACTTTAATAAGCCCCATAACTTACCTGATCTCGACGACACCTATACTAAATTCTTAA
Protein
MLDQKIIQPSLSPWSSPIWIVPKKTDASGQKKWRIVIDYRKLNDITIGETYPIPNIVEILDQLGNSKYFTTLDLASGFHQITMNQKDACKTAFSVPQGHFEFTRMPFGLKNAPSTFQRLMNNVLTGLQGERCFVYLDDIVSYSHDLKTHIENLGAIFDRLRKFNLKLQPDKCEFLRKEVGYLGHIISEEGLKPDPNKIKSVKEFPIPKSPKDIKSFLGLISYYRRFIPEFSKLSKPLTSLLKKDASFLWTNEQQLAFETLKDKLITSPVLIYPDFTKPFNLTCDASNYAISAILSQGPIGKDRPVAYASRTLNKCEINYSTTEKELLAIVWGCNSFRPYLFGRKFIIVTDHRPLKWLFNHTDPSSKLQRWRLQLQEFEYEIIYRKGKLNSAADALSRYPVNPVYTENPENLNENELIDLGPLDISPFNDEVPDFTPLSPSPNNLPDTLLQPEIVPEITDQNPIIENEPQPSSSQAPILQQPNTLISPITYLISTTPILNS
Summary
Catalytic Activity
a 2'-deoxyribonucleoside 5'-triphosphate + DNA(n) = diphosphate + DNA(n+1)
Miscellaneous
The open reading frame is located in a copia-like transposable element called 17.6.
Keywords
Aspartyl protease
Endonuclease
Hydrolase
Nuclease
Nucleotidyltransferase
Protease
RNA-directed DNA polymerase
Transferase
Transposable element
Feature
chain Retrovirus-related Pol polyprotein from transposon 17.6
Uniprot
E0VZF9
E0VZF7
A0A1Y1KJL2
D7GXP0
A0A0J7KFZ4
A0A0J7K6M0
+ More
A0A2J7PEY0 A0A2J7RJQ6 J9LRA5 A0A2S2NLS2 D7EKG9 A0A2J7QWM2 A0A2J7Q0T0 A0A0J7JY03 A0A2J7R948 A0A2S2NNZ3 A0A2J7PPW9 A0A2S2NN14 A0A2S2QW04 A0A2J7PP03 A0A0K8TSF6 A0A2J7RBZ9 A0A0K8TRU1 A0A2J7QTC1 X1WUD2 Q65353 A0A0J7KIJ2 X1WM57 A0A2J7PFW5 X1WLD0 A0A3Q0JL77 A0A0J7N7Q7 X1X2K6 A0A0J7KM44 A0A1Y1KML2 A0A2J7RRF2 A0A2J7PDA1 J9M441 A0A355BCA1 A0A0J7JZT1 A0A2J7PRU8 A0A0A9Z1T0 A0A0A9XJZ5 A0A2S2P749 A0A1B6DI21 Q5BN17 A0A2S2NFT1 U5ES61 X1WLD3 A0A1B6E8Y4 A0A2H8TVW1 X1WLJ7 A0A1W7R6F0 V5GQE1 A0A224XA16 A0A0J7KIE0 A0A2H8THY5 A0A3Q0JCA5 A0A1W7R695 A0A3Q0JG65 A0A1W7R6J9 V5I864 O46115 X1XQI4 V5FY26 D6WE31 A0A224XIQ6 A0A2S2NQ60 A0A0A9XD39 A0A0A9XG63 J9LWC7 A0A2S2PBQ7 Q967T2 A0A146L836 A0A0K8WF08 X1X3W2 J9KPM2 K7JCQ6 A0A1W7R686 O96740 O76326 J9LJV3 A0A0K8WBI5 A0A2S2PWE1 J9LS83 P04323 K7IUU7 A0A0A9YU38 A0A146KMA2 J9KGC3 X1WUN1 A0A0K8S7R8 Q8T5U3 A0A0A9YJ44 X1WX89
A0A2J7PEY0 A0A2J7RJQ6 J9LRA5 A0A2S2NLS2 D7EKG9 A0A2J7QWM2 A0A2J7Q0T0 A0A0J7JY03 A0A2J7R948 A0A2S2NNZ3 A0A2J7PPW9 A0A2S2NN14 A0A2S2QW04 A0A2J7PP03 A0A0K8TSF6 A0A2J7RBZ9 A0A0K8TRU1 A0A2J7QTC1 X1WUD2 Q65353 A0A0J7KIJ2 X1WM57 A0A2J7PFW5 X1WLD0 A0A3Q0JL77 A0A0J7N7Q7 X1X2K6 A0A0J7KM44 A0A1Y1KML2 A0A2J7RRF2 A0A2J7PDA1 J9M441 A0A355BCA1 A0A0J7JZT1 A0A2J7PRU8 A0A0A9Z1T0 A0A0A9XJZ5 A0A2S2P749 A0A1B6DI21 Q5BN17 A0A2S2NFT1 U5ES61 X1WLD3 A0A1B6E8Y4 A0A2H8TVW1 X1WLJ7 A0A1W7R6F0 V5GQE1 A0A224XA16 A0A0J7KIE0 A0A2H8THY5 A0A3Q0JCA5 A0A1W7R695 A0A3Q0JG65 A0A1W7R6J9 V5I864 O46115 X1XQI4 V5FY26 D6WE31 A0A224XIQ6 A0A2S2NQ60 A0A0A9XD39 A0A0A9XG63 J9LWC7 A0A2S2PBQ7 Q967T2 A0A146L836 A0A0K8WF08 X1X3W2 J9KPM2 K7JCQ6 A0A1W7R686 O96740 O76326 J9LJV3 A0A0K8WBI5 A0A2S2PWE1 J9LS83 P04323 K7IUU7 A0A0A9YU38 A0A146KMA2 J9KGC3 X1WUN1 A0A0K8S7R8 Q8T5U3 A0A0A9YJ44 X1WX89
Pubmed
EMBL
DS235852
EEB18765.1
EEB18763.1
GEZM01081533
JAV61609.1
GG694348
+ More
EFA13518.1 LBMM01008064 KMQ89174.1 LBMM01013045 KMQ85846.1 NEVH01026087 PNF14896.1 NEVH01002988 PNF41072.1 ABLF02030471 ABLF02066562 GGMR01005468 MBY18087.1 DS497745 EFA13136.1 NEVH01009425 PNF32982.1 NEVH01019964 PNF22169.1 LBMM01022303 KMQ82914.1 NEVH01006666 PNF37360.1 GGMR01006290 MBY18909.1 NEVH01022642 PNF18384.1 GGMR01005970 MBY18589.1 GGMS01012487 MBY81690.1 NEVH01023280 PNF18074.1 GDAI01000738 JAI16865.1 NEVH01005888 PNF38366.1 GDAI01000740 JAI16863.1 NEVH01011195 PNF31840.1 ABLF02041284 M32662 AAA92249.1 LBMM01007099 KMQ90041.1 ABLF02001345 ABLF02006651 ABLF02006655 ABLF02054079 ABLF02054906 ABLF02062695 NEVH01025646 PNF15218.1 ABLF02041728 LBMM01008712 KMQ88680.1 ABLF02024213 ABLF02042472 LBMM01005670 LBMM01005669 KMQ91307.1 KMQ91309.1 GEZM01078735 JAV62639.1 NEVH01000607 PNF43410.1 NEVH01026392 PNF14303.1 DNYX01000282 HBK83635.1 LBMM01019926 KMQ83421.1 NEVH01021952 PNF19057.1 GBHO01004397 JAG39207.1 GBHO01026164 JAG17440.1 GGMR01012631 MBY25250.1 GEDC01017847 GEDC01011974 JAS19451.1 JAS25324.1 AY928610 AAX28844.1 GGMR01003444 MBY16063.1 GANO01002504 JAB57367.1 ABLF02011604 ABLF02041632 GEDC01002915 JAS34383.1 GFXV01006571 MBW18376.1 ABLF02011176 ABLF02041729 GEHC01000906 JAV46739.1 GALX01004629 JAB63837.1 GFTR01008672 JAW07754.1 LBMM01006919 LBMM01006918 KMQ90193.1 KMQ90194.1 GFXV01001507 MBW13312.1 GEHC01000948 JAV46697.1 GEHC01000905 JAV46740.1 GALX01005230 JAB63236.1 AJ000387 CAA04050.1 ABLF02041376 GALX01005891 JAB62575.1 KQ971324 EFA01226.1 GFTR01008505 JAW07921.1 GGMR01006639 MBY19258.1 GBHO01024992 JAG18612.1 GBHO01024993 JAG18611.1 ABLF02012660 ABLF02012662 GGMR01014274 MBY26893.1 AF364550 AAK52058.1 GDHC01015253 JAQ03376.1 GDHF01002622 JAI49692.1 ABLF02041371 AAZX01005108 GEHC01000961 JAV46684.1 AJ009736 CAA08807.1 AF056940 AAC33318.1 ABLF02008732 GDHF01004109 JAI48205.1 GGMS01000510 MBY69713.1 X01472 AAZX01005109 GBHO01007885 JAG35719.1 GDHC01021754 JAP96874.1 ABLF02065961 ABLF02025379 ABLF02025385 ABLF02041365 ABLF02057103 GBRD01016627 JAG49199.1 AF492764 AAM11674.1 GBHO01012506 JAG31098.1 ABLF02035279 ABLF02041373
EFA13518.1 LBMM01008064 KMQ89174.1 LBMM01013045 KMQ85846.1 NEVH01026087 PNF14896.1 NEVH01002988 PNF41072.1 ABLF02030471 ABLF02066562 GGMR01005468 MBY18087.1 DS497745 EFA13136.1 NEVH01009425 PNF32982.1 NEVH01019964 PNF22169.1 LBMM01022303 KMQ82914.1 NEVH01006666 PNF37360.1 GGMR01006290 MBY18909.1 NEVH01022642 PNF18384.1 GGMR01005970 MBY18589.1 GGMS01012487 MBY81690.1 NEVH01023280 PNF18074.1 GDAI01000738 JAI16865.1 NEVH01005888 PNF38366.1 GDAI01000740 JAI16863.1 NEVH01011195 PNF31840.1 ABLF02041284 M32662 AAA92249.1 LBMM01007099 KMQ90041.1 ABLF02001345 ABLF02006651 ABLF02006655 ABLF02054079 ABLF02054906 ABLF02062695 NEVH01025646 PNF15218.1 ABLF02041728 LBMM01008712 KMQ88680.1 ABLF02024213 ABLF02042472 LBMM01005670 LBMM01005669 KMQ91307.1 KMQ91309.1 GEZM01078735 JAV62639.1 NEVH01000607 PNF43410.1 NEVH01026392 PNF14303.1 DNYX01000282 HBK83635.1 LBMM01019926 KMQ83421.1 NEVH01021952 PNF19057.1 GBHO01004397 JAG39207.1 GBHO01026164 JAG17440.1 GGMR01012631 MBY25250.1 GEDC01017847 GEDC01011974 JAS19451.1 JAS25324.1 AY928610 AAX28844.1 GGMR01003444 MBY16063.1 GANO01002504 JAB57367.1 ABLF02011604 ABLF02041632 GEDC01002915 JAS34383.1 GFXV01006571 MBW18376.1 ABLF02011176 ABLF02041729 GEHC01000906 JAV46739.1 GALX01004629 JAB63837.1 GFTR01008672 JAW07754.1 LBMM01006919 LBMM01006918 KMQ90193.1 KMQ90194.1 GFXV01001507 MBW13312.1 GEHC01000948 JAV46697.1 GEHC01000905 JAV46740.1 GALX01005230 JAB63236.1 AJ000387 CAA04050.1 ABLF02041376 GALX01005891 JAB62575.1 KQ971324 EFA01226.1 GFTR01008505 JAW07921.1 GGMR01006639 MBY19258.1 GBHO01024992 JAG18612.1 GBHO01024993 JAG18611.1 ABLF02012660 ABLF02012662 GGMR01014274 MBY26893.1 AF364550 AAK52058.1 GDHC01015253 JAQ03376.1 GDHF01002622 JAI49692.1 ABLF02041371 AAZX01005108 GEHC01000961 JAV46684.1 AJ009736 CAA08807.1 AF056940 AAC33318.1 ABLF02008732 GDHF01004109 JAI48205.1 GGMS01000510 MBY69713.1 X01472 AAZX01005109 GBHO01007885 JAG35719.1 GDHC01021754 JAP96874.1 ABLF02065961 ABLF02025379 ABLF02025385 ABLF02041365 ABLF02057103 GBRD01016627 JAG49199.1 AF492764 AAM11674.1 GBHO01012506 JAG31098.1 ABLF02035279 ABLF02041373
Proteomes
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR041588 Integrase_H2C2
IPR000477 RT_dom
IPR041373 RT_RNaseH
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR002589 Macro_dom
IPR018061 Retropepsins
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR022048 Envelope_fusion-like
IPR003653 Peptidase_C48_C
IPR038765 Papain-like_cys_pep_sf
IPR005162 Retrotrans_gag_dom
IPR041588 Integrase_H2C2
IPR000477 RT_dom
IPR041373 RT_RNaseH
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR021109 Peptidase_aspartic_dom_sf
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR002589 Macro_dom
IPR018061 Retropepsins
IPR036875 Znf_CCHC_sf
IPR001878 Znf_CCHC
IPR022048 Envelope_fusion-like
IPR003653 Peptidase_C48_C
IPR038765 Papain-like_cys_pep_sf
IPR005162 Retrotrans_gag_dom
Gene 3D
ProteinModelPortal
E0VZF9
E0VZF7
A0A1Y1KJL2
D7GXP0
A0A0J7KFZ4
A0A0J7K6M0
+ More
A0A2J7PEY0 A0A2J7RJQ6 J9LRA5 A0A2S2NLS2 D7EKG9 A0A2J7QWM2 A0A2J7Q0T0 A0A0J7JY03 A0A2J7R948 A0A2S2NNZ3 A0A2J7PPW9 A0A2S2NN14 A0A2S2QW04 A0A2J7PP03 A0A0K8TSF6 A0A2J7RBZ9 A0A0K8TRU1 A0A2J7QTC1 X1WUD2 Q65353 A0A0J7KIJ2 X1WM57 A0A2J7PFW5 X1WLD0 A0A3Q0JL77 A0A0J7N7Q7 X1X2K6 A0A0J7KM44 A0A1Y1KML2 A0A2J7RRF2 A0A2J7PDA1 J9M441 A0A355BCA1 A0A0J7JZT1 A0A2J7PRU8 A0A0A9Z1T0 A0A0A9XJZ5 A0A2S2P749 A0A1B6DI21 Q5BN17 A0A2S2NFT1 U5ES61 X1WLD3 A0A1B6E8Y4 A0A2H8TVW1 X1WLJ7 A0A1W7R6F0 V5GQE1 A0A224XA16 A0A0J7KIE0 A0A2H8THY5 A0A3Q0JCA5 A0A1W7R695 A0A3Q0JG65 A0A1W7R6J9 V5I864 O46115 X1XQI4 V5FY26 D6WE31 A0A224XIQ6 A0A2S2NQ60 A0A0A9XD39 A0A0A9XG63 J9LWC7 A0A2S2PBQ7 Q967T2 A0A146L836 A0A0K8WF08 X1X3W2 J9KPM2 K7JCQ6 A0A1W7R686 O96740 O76326 J9LJV3 A0A0K8WBI5 A0A2S2PWE1 J9LS83 P04323 K7IUU7 A0A0A9YU38 A0A146KMA2 J9KGC3 X1WUN1 A0A0K8S7R8 Q8T5U3 A0A0A9YJ44 X1WX89
A0A2J7PEY0 A0A2J7RJQ6 J9LRA5 A0A2S2NLS2 D7EKG9 A0A2J7QWM2 A0A2J7Q0T0 A0A0J7JY03 A0A2J7R948 A0A2S2NNZ3 A0A2J7PPW9 A0A2S2NN14 A0A2S2QW04 A0A2J7PP03 A0A0K8TSF6 A0A2J7RBZ9 A0A0K8TRU1 A0A2J7QTC1 X1WUD2 Q65353 A0A0J7KIJ2 X1WM57 A0A2J7PFW5 X1WLD0 A0A3Q0JL77 A0A0J7N7Q7 X1X2K6 A0A0J7KM44 A0A1Y1KML2 A0A2J7RRF2 A0A2J7PDA1 J9M441 A0A355BCA1 A0A0J7JZT1 A0A2J7PRU8 A0A0A9Z1T0 A0A0A9XJZ5 A0A2S2P749 A0A1B6DI21 Q5BN17 A0A2S2NFT1 U5ES61 X1WLD3 A0A1B6E8Y4 A0A2H8TVW1 X1WLJ7 A0A1W7R6F0 V5GQE1 A0A224XA16 A0A0J7KIE0 A0A2H8THY5 A0A3Q0JCA5 A0A1W7R695 A0A3Q0JG65 A0A1W7R6J9 V5I864 O46115 X1XQI4 V5FY26 D6WE31 A0A224XIQ6 A0A2S2NQ60 A0A0A9XD39 A0A0A9XG63 J9LWC7 A0A2S2PBQ7 Q967T2 A0A146L836 A0A0K8WF08 X1X3W2 J9KPM2 K7JCQ6 A0A1W7R686 O96740 O76326 J9LJV3 A0A0K8WBI5 A0A2S2PWE1 J9LS83 P04323 K7IUU7 A0A0A9YU38 A0A146KMA2 J9KGC3 X1WUN1 A0A0K8S7R8 Q8T5U3 A0A0A9YJ44 X1WX89
PDB
4OL8
E-value=7.118e-65,
Score=628
Ontologies
GO
Topology
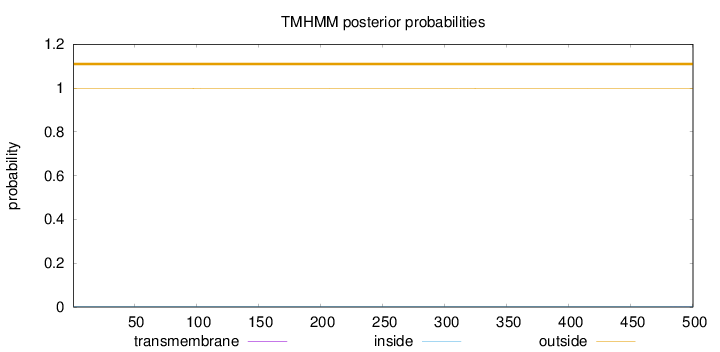
Length:
500
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00385
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.00177
outside
1 - 500
Population Genetic Test Statistics
Pi
163.326149
Theta
64.917587
Tajima's D
4.73293
CLR
0
CSRT
0.999950002499875
Interpretation
Uncertain
