Gene
KWMTBOMO10797
Pre Gene Modal
BGIBMGA008436
Annotation
PREDICTED:_disheveled-associated_activator_of_morphogenesis_1_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.405
Sequence
CDS
ATGGCGCCCGGGAAGCTGGTTCACGCATTATCGCTTAGTCACATATACGATCCTGACGTGGCTCCCCTCGAGATAAACGTCGGCGAGATAGTGCACCTGCTCGCCAAGGAGGAAGAGCTCGTCGCCGCCAGAACCAAGGCCGAGAATCTGGAACGGGAGAACATCGACCTGGCCACAGAGCTGGCTAAGAAGGAGAAGCAGCTGGACGAGCGCGCGCAGGAGCGCGAGGAGCTGGAGGGCGCCGCGGCGCGCCTGCGGGACCGCCTGGAGCGCGAGACGGCCGCGCACATGCACTGCGCCGACCTGGCGCGCGCCGCCGCCGCACGCGCGCAGCTGCTCGACTGCCAGTTGACCCAAGAGAGAACCGAGAGAGCGAGATTCGAAAAACTAGTCAGCGAGGGAAGCATACCTGATGATGCTAAGGTGAACAACCTCAAGAACGCGAGCTTCAACTGGAGTAAACTACCTGATACCAAGCTGCACGGCACGATATGGCAGGAGCTAGACGACACGAAGCTGTACAACGCGATGGATCTGCACACCATCGACAAGATGTTCTGTGCCTACCAGAAGAATGGCGTCCAGAACGAGGGGTCCGTCGAGGACCTCCGCCAGCTCGGCTCGAAGCCCAGGACGAAGATACTGTCCGTGATAGACGGCCGCCGCGCCCAGAACTGCACCATCCTGCTGTCCAAACTTAAAATGACCGATGAGGAGATTTGCAGAGCGATCCTCAAAATGGACAGCGGTGAACAACTGCCCATCGATATGTTGGAGCAACTCCTGAAGTTCACGCCCAGCGCCGAGGAGGCCGCCATGCTAGAAGAGCACCAAGATGAACTGGACAGCATGGCGAGGGCGGATCGCTTCCTCTACGAGATCTCCAAGATCCCGCACTACTCGATGCGGGTGCGCACGCTGCTGTTCAAGAAGCGGTTCGCGGCGGCGAGCGCGGAGGCGAGCGGGCGCGCCACCACCGTGCTGCGCGCCGCGCGGGACATGACGCGCTCGCGCCGCCTGCGCGCGCTGCTCGAGCTGGTGCTGGCGCTCGGGAACTACATGAACAGGGGAGCCCGCGGCAACGCGTCAGGGTTCCGGCTCTCGTCACTGAACAAACTCGCCGACACCAAGTCCAGTGTTACAAGAAACACCACGCTCCTGCATTTCCTGGTCGAAATGTTGGAGACACAGTTCAAAGACATACTGCTGCTCGAAGAGGATTTACCGCACGTCCGAGCCGCGGCCAAGGTTTGCGTGGAACAACTGGAGAAAGATGTGGGTGCTTTGAGGAGTGGTCTCCGTGAAGTCGCGAAGGAGGTGGAGTACCACGCGTCGCTGCCCTCACCGCAGCCCGGTGACGCCTTCCTACCCGTCATGAGGGAGTTCCATGCGCATGCCGTTTGTACCTTCACACAACTGGAAGACCTTTTCCAGGACATGAAGAGTCGTCTCGAAGCATGCGCCCACGCCTTCGGTGAAGAGACAAGTGCGTCCCCGGAGCAACTGTTCGGCGCCATGGACGCGTTCCTCACGCAGCTTGCGGAGGCGCGCGCCGAGTGCGACGCCATCAGGAGGCGCCGCGACGACGAGCAGCGCCGGACCAGGCATGAGCAAGAGTTGAAAAAGCGTACAATGGAACGTAAGCAATCAAACTCGCTAGGCTCGGTGAGCAAATCCCTAGCGAAATCCAACGGGGATTGCAACGGACACTCCAACGATAGCTCCCGGGACGGCACCATGAGTAACGGACAAAAAGGAGAGTTCGATGACCTTATATCCGCATTACGAACAGGAGATGTCTTCGGAGATGACGTGGCCAAATTCAAAAGATCAAGAAAGACTTCCAAAGTCACTCAGAAGGGTCGCGACTCTCCTCCCAGGGGCGTGTGCAGGGAAGACTCTAGGGAGAGACAGAAGAATTGA
Protein
MAPGKLVHALSLSHIYDPDVAPLEINVGEIVHLLAKEEELVAARTKAENLERENIDLATELAKKEKQLDERAQEREELEGAAARLRDRLERETAAHMHCADLARAAAARAQLLDCQLTQERTERARFEKLVSEGSIPDDAKVNNLKNASFNWSKLPDTKLHGTIWQELDDTKLYNAMDLHTIDKMFCAYQKNGVQNEGSVEDLRQLGSKPRTKILSVIDGRRAQNCTILLSKLKMTDEEICRAILKMDSGEQLPIDMLEQLLKFTPSAEEAAMLEEHQDELDSMARADRFLYEISKIPHYSMRVRTLLFKKRFAAASAEASGRATTVLRAARDMTRSRRLRALLELVLALGNYMNRGARGNASGFRLSSLNKLADTKSSVTRNTTLLHFLVEMLETQFKDILLLEEDLPHVRAAAKVCVEQLEKDVGALRSGLREVAKEVEYHASLPSPQPGDAFLPVMREFHAHAVCTFTQLEDLFQDMKSRLEACAHAFGEETSASPEQLFGAMDAFLTQLAEARAECDAIRRRRDDEQRRTRHEQELKKRTMERKQSNSLGSVSKSLAKSNGDCNGHSNDSSRDGTMSNGQKGEFDDLISALRTGDVFGDDVAKFKRSRKTSKVTQKGRDSPPRGVCREDSRERQKN
Summary
Uniprot
A0A194PE00
H9JFY9
A0A2H1W0G9
A0A1W4WX95
A0A1W4WL92
A0A1W4WVZ1
+ More
A0A1W4WX10 D2A1M1 A0A067R5U9 Q7QKJ3 A0A1Y1LB84 A0A154P3Z0 A0A182GZE7 Q16HS4 E2BJR3 A0A2C9H3V9 A0A182P5R7 A0A087ZVL2 A0A182FQP8 A0A182NL63 W5J1H0 A0A0L7RAH1 E2AS93 A0A1S4G7X6 A0A195CAC0 A0A182VQM0 A0A310SE13 A0A182JSN3 A0A182RHX9 A0A182UDJ2 F4WUD6 A0A026W786 A0A0M8ZWQ7 A0A151WUC1 A0A195FDI1 A0A151J034 A0A182MR58 A0A182LCR6 A0A1B6CJL8 A0A182YK93 A0A182QN43 A0A182II20 A0A182IWE4 A0A1B6D4F8 A0A1B6C417 A0A084VQL4 A0A158NIU2 A0A182UWJ5 A0A0C9QEL2 A0A195BZC4 A0A336LG97 A0A336MYR9 A0A336N1D8 A0A0C9R184 A0A0C9R4G2 A0A0A9X1K2 W8ALP6 A0A1A9VCS2 A0A0A1XAP4 A0A1Q3G3F8 A0A0K8VN44 A0A1I8P4R0 A0A1I8NCD6 A0A1A9XBL5 A0A034VK79 W8AQT8 T1PIB8 A0A1A9WCN2 A0A2J7PXE8 W8BAK2 A0A0V0G703 A0A1A9ZER4 A0A034VHH4 A0A1Q3G3N0 A0A023F3Z6 A0A1Q3G3I4 A0A034VJH9 A0A1Q3G2V5 A0A1B0ATF1 A0A224XAB6 T1HSY1 A0A069DX58 A0A0L0CRM7 A0A0P4VZV7 E0VIP0 Q7YZA4 A0A0Q5ST07 A0A1B6IC32 K7J3N6 Q8IRY0 A0A146MFK9 Q8MT16 A0A146LF02 A0A146LBJ9 A0A1B0CAZ8 B4I9C0 B3P9G0 A0A0Q5SU66
A0A1W4WX10 D2A1M1 A0A067R5U9 Q7QKJ3 A0A1Y1LB84 A0A154P3Z0 A0A182GZE7 Q16HS4 E2BJR3 A0A2C9H3V9 A0A182P5R7 A0A087ZVL2 A0A182FQP8 A0A182NL63 W5J1H0 A0A0L7RAH1 E2AS93 A0A1S4G7X6 A0A195CAC0 A0A182VQM0 A0A310SE13 A0A182JSN3 A0A182RHX9 A0A182UDJ2 F4WUD6 A0A026W786 A0A0M8ZWQ7 A0A151WUC1 A0A195FDI1 A0A151J034 A0A182MR58 A0A182LCR6 A0A1B6CJL8 A0A182YK93 A0A182QN43 A0A182II20 A0A182IWE4 A0A1B6D4F8 A0A1B6C417 A0A084VQL4 A0A158NIU2 A0A182UWJ5 A0A0C9QEL2 A0A195BZC4 A0A336LG97 A0A336MYR9 A0A336N1D8 A0A0C9R184 A0A0C9R4G2 A0A0A9X1K2 W8ALP6 A0A1A9VCS2 A0A0A1XAP4 A0A1Q3G3F8 A0A0K8VN44 A0A1I8P4R0 A0A1I8NCD6 A0A1A9XBL5 A0A034VK79 W8AQT8 T1PIB8 A0A1A9WCN2 A0A2J7PXE8 W8BAK2 A0A0V0G703 A0A1A9ZER4 A0A034VHH4 A0A1Q3G3N0 A0A023F3Z6 A0A1Q3G3I4 A0A034VJH9 A0A1Q3G2V5 A0A1B0ATF1 A0A224XAB6 T1HSY1 A0A069DX58 A0A0L0CRM7 A0A0P4VZV7 E0VIP0 Q7YZA4 A0A0Q5ST07 A0A1B6IC32 K7J3N6 Q8IRY0 A0A146MFK9 Q8MT16 A0A146LF02 A0A146LBJ9 A0A1B0CAZ8 B4I9C0 B3P9G0 A0A0Q5SU66
Pubmed
26354079
19121390
18362917
19820115
24845553
12364791
+ More
28004739 26483478 17510324 20798317 20920257 23761445 21719571 24508170 30249741 20966253 25244985 24438588 21347285 25401762 24495485 25830018 25315136 25348373 25474469 26334808 26108605 27129103 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 18057021 20075255 26109357 26109356 26823975
28004739 26483478 17510324 20798317 20920257 23761445 21719571 24508170 30249741 20966253 25244985 24438588 21347285 25401762 24495485 25830018 25315136 25348373 25474469 26334808 26108605 27129103 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 18057021 20075255 26109357 26109356 26823975
EMBL
KQ459606
KPI90914.1
BABH01001621
BABH01001622
BABH01001623
BABH01001624
+ More
BABH01001625 ODYU01005559 SOQ46527.1 KQ971338 EFA01522.1 KK852867 KDR14722.1 AAAB01008794 EAA03583.5 GEZM01063211 JAV69195.1 KQ434804 KZC06054.1 JXUM01099401 KQ564489 KXJ72200.1 CH478145 EAT33802.1 GL448616 EFN84071.1 ADMH02002181 ETN58017.1 KQ414618 KOC67835.1 GL442229 EFN63723.1 KQ978068 KYM97635.1 KQ766789 OAD53573.1 GL888356 EGI62191.1 KK107432 QOIP01000001 EZA50889.1 RLU26859.1 KQ435845 KOX71173.1 KQ982730 KYQ51519.1 KQ981677 KYN38104.1 KQ980665 KYN14644.1 AXCM01001179 GEDC01023688 JAS13610.1 AXCN02000083 APCN01002159 APCN01002160 GEDC01016709 GEDC01013978 JAS20589.1 JAS23320.1 GEDC01029052 JAS08246.1 ATLV01015250 KE525004 KFB40258.1 ADTU01017170 ADTU01017171 ADTU01017172 ADTU01017173 GBYB01001714 JAG71481.1 KQ976394 KYM93268.1 UFQS01003390 UFQT01003390 SSX15595.1 SSX34957.1 UFQT01003906 SSX35360.1 SSX35359.1 GBYB01001720 JAG71487.1 GBYB01001716 JAG71483.1 GBHO01030981 JAG12623.1 GAMC01019688 JAB86867.1 GBXI01006539 JAD07753.1 GFDL01000708 JAV34337.1 GDHF01018434 GDHF01012012 JAI33880.1 JAI40302.1 GAKP01016984 JAC41968.1 GAMC01019687 JAB86868.1 KA647880 AFP62509.1 NEVH01020859 PNF21002.1 GAMC01019686 JAB86869.1 GECL01002334 JAP03790.1 GAKP01016985 GAKP01016983 JAC41969.1 GFDL01000676 JAV34369.1 GBBI01002551 JAC16161.1 GFDL01000685 JAV34360.1 GAKP01016982 JAC41970.1 GFDL01000898 JAV34147.1 JXJN01003291 GFTR01008562 JAW07864.1 ACPB03016857 ACPB03016858 GBGD01000226 JAC88663.1 JRES01000104 KNC34099.1 GDKW01001533 JAI55062.1 DS235201 EEB13246.1 AE014298 AAF45600.2 CH954183 KQS25966.1 KQS25967.1 GECU01023192 JAS84514.1 AAF45601.3 AAN09040.1 AGB94972.1 AHN59242.1 GDHC01000654 JAQ17975.1 AY118439 AAM48468.1 GDHC01011888 JAQ06741.1 GDHC01014182 JAQ04447.1 AJWK01004687 AJWK01004688 CH480825 EDW43801.1 EDV45456.2 KQS25968.1
BABH01001625 ODYU01005559 SOQ46527.1 KQ971338 EFA01522.1 KK852867 KDR14722.1 AAAB01008794 EAA03583.5 GEZM01063211 JAV69195.1 KQ434804 KZC06054.1 JXUM01099401 KQ564489 KXJ72200.1 CH478145 EAT33802.1 GL448616 EFN84071.1 ADMH02002181 ETN58017.1 KQ414618 KOC67835.1 GL442229 EFN63723.1 KQ978068 KYM97635.1 KQ766789 OAD53573.1 GL888356 EGI62191.1 KK107432 QOIP01000001 EZA50889.1 RLU26859.1 KQ435845 KOX71173.1 KQ982730 KYQ51519.1 KQ981677 KYN38104.1 KQ980665 KYN14644.1 AXCM01001179 GEDC01023688 JAS13610.1 AXCN02000083 APCN01002159 APCN01002160 GEDC01016709 GEDC01013978 JAS20589.1 JAS23320.1 GEDC01029052 JAS08246.1 ATLV01015250 KE525004 KFB40258.1 ADTU01017170 ADTU01017171 ADTU01017172 ADTU01017173 GBYB01001714 JAG71481.1 KQ976394 KYM93268.1 UFQS01003390 UFQT01003390 SSX15595.1 SSX34957.1 UFQT01003906 SSX35360.1 SSX35359.1 GBYB01001720 JAG71487.1 GBYB01001716 JAG71483.1 GBHO01030981 JAG12623.1 GAMC01019688 JAB86867.1 GBXI01006539 JAD07753.1 GFDL01000708 JAV34337.1 GDHF01018434 GDHF01012012 JAI33880.1 JAI40302.1 GAKP01016984 JAC41968.1 GAMC01019687 JAB86868.1 KA647880 AFP62509.1 NEVH01020859 PNF21002.1 GAMC01019686 JAB86869.1 GECL01002334 JAP03790.1 GAKP01016985 GAKP01016983 JAC41969.1 GFDL01000676 JAV34369.1 GBBI01002551 JAC16161.1 GFDL01000685 JAV34360.1 GAKP01016982 JAC41970.1 GFDL01000898 JAV34147.1 JXJN01003291 GFTR01008562 JAW07864.1 ACPB03016857 ACPB03016858 GBGD01000226 JAC88663.1 JRES01000104 KNC34099.1 GDKW01001533 JAI55062.1 DS235201 EEB13246.1 AE014298 AAF45600.2 CH954183 KQS25966.1 KQS25967.1 GECU01023192 JAS84514.1 AAF45601.3 AAN09040.1 AGB94972.1 AHN59242.1 GDHC01000654 JAQ17975.1 AY118439 AAM48468.1 GDHC01011888 JAQ06741.1 GDHC01014182 JAQ04447.1 AJWK01004687 AJWK01004688 CH480825 EDW43801.1 EDV45456.2 KQS25968.1
Proteomes
UP000053268
UP000005204
UP000192223
UP000007266
UP000027135
UP000007062
+ More
UP000076502 UP000069940 UP000249989 UP000008820 UP000008237 UP000075885 UP000005203 UP000069272 UP000075884 UP000000673 UP000053825 UP000000311 UP000078542 UP000075920 UP000075881 UP000075900 UP000075902 UP000007755 UP000053097 UP000279307 UP000053105 UP000075809 UP000078541 UP000078492 UP000075883 UP000075882 UP000076408 UP000075886 UP000075840 UP000075880 UP000030765 UP000005205 UP000075903 UP000078540 UP000078200 UP000095300 UP000095301 UP000092443 UP000091820 UP000235965 UP000092445 UP000092460 UP000015103 UP000037069 UP000009046 UP000000803 UP000008711 UP000002358 UP000092461 UP000001292
UP000076502 UP000069940 UP000249989 UP000008820 UP000008237 UP000075885 UP000005203 UP000069272 UP000075884 UP000000673 UP000053825 UP000000311 UP000078542 UP000075920 UP000075881 UP000075900 UP000075902 UP000007755 UP000053097 UP000279307 UP000053105 UP000075809 UP000078541 UP000078492 UP000075883 UP000075882 UP000076408 UP000075886 UP000075840 UP000075880 UP000030765 UP000005205 UP000075903 UP000078540 UP000078200 UP000095300 UP000095301 UP000092443 UP000091820 UP000235965 UP000092445 UP000092460 UP000015103 UP000037069 UP000009046 UP000000803 UP000008711 UP000002358 UP000092461 UP000001292
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A194PE00
H9JFY9
A0A2H1W0G9
A0A1W4WX95
A0A1W4WL92
A0A1W4WVZ1
+ More
A0A1W4WX10 D2A1M1 A0A067R5U9 Q7QKJ3 A0A1Y1LB84 A0A154P3Z0 A0A182GZE7 Q16HS4 E2BJR3 A0A2C9H3V9 A0A182P5R7 A0A087ZVL2 A0A182FQP8 A0A182NL63 W5J1H0 A0A0L7RAH1 E2AS93 A0A1S4G7X6 A0A195CAC0 A0A182VQM0 A0A310SE13 A0A182JSN3 A0A182RHX9 A0A182UDJ2 F4WUD6 A0A026W786 A0A0M8ZWQ7 A0A151WUC1 A0A195FDI1 A0A151J034 A0A182MR58 A0A182LCR6 A0A1B6CJL8 A0A182YK93 A0A182QN43 A0A182II20 A0A182IWE4 A0A1B6D4F8 A0A1B6C417 A0A084VQL4 A0A158NIU2 A0A182UWJ5 A0A0C9QEL2 A0A195BZC4 A0A336LG97 A0A336MYR9 A0A336N1D8 A0A0C9R184 A0A0C9R4G2 A0A0A9X1K2 W8ALP6 A0A1A9VCS2 A0A0A1XAP4 A0A1Q3G3F8 A0A0K8VN44 A0A1I8P4R0 A0A1I8NCD6 A0A1A9XBL5 A0A034VK79 W8AQT8 T1PIB8 A0A1A9WCN2 A0A2J7PXE8 W8BAK2 A0A0V0G703 A0A1A9ZER4 A0A034VHH4 A0A1Q3G3N0 A0A023F3Z6 A0A1Q3G3I4 A0A034VJH9 A0A1Q3G2V5 A0A1B0ATF1 A0A224XAB6 T1HSY1 A0A069DX58 A0A0L0CRM7 A0A0P4VZV7 E0VIP0 Q7YZA4 A0A0Q5ST07 A0A1B6IC32 K7J3N6 Q8IRY0 A0A146MFK9 Q8MT16 A0A146LF02 A0A146LBJ9 A0A1B0CAZ8 B4I9C0 B3P9G0 A0A0Q5SU66
A0A1W4WX10 D2A1M1 A0A067R5U9 Q7QKJ3 A0A1Y1LB84 A0A154P3Z0 A0A182GZE7 Q16HS4 E2BJR3 A0A2C9H3V9 A0A182P5R7 A0A087ZVL2 A0A182FQP8 A0A182NL63 W5J1H0 A0A0L7RAH1 E2AS93 A0A1S4G7X6 A0A195CAC0 A0A182VQM0 A0A310SE13 A0A182JSN3 A0A182RHX9 A0A182UDJ2 F4WUD6 A0A026W786 A0A0M8ZWQ7 A0A151WUC1 A0A195FDI1 A0A151J034 A0A182MR58 A0A182LCR6 A0A1B6CJL8 A0A182YK93 A0A182QN43 A0A182II20 A0A182IWE4 A0A1B6D4F8 A0A1B6C417 A0A084VQL4 A0A158NIU2 A0A182UWJ5 A0A0C9QEL2 A0A195BZC4 A0A336LG97 A0A336MYR9 A0A336N1D8 A0A0C9R184 A0A0C9R4G2 A0A0A9X1K2 W8ALP6 A0A1A9VCS2 A0A0A1XAP4 A0A1Q3G3F8 A0A0K8VN44 A0A1I8P4R0 A0A1I8NCD6 A0A1A9XBL5 A0A034VK79 W8AQT8 T1PIB8 A0A1A9WCN2 A0A2J7PXE8 W8BAK2 A0A0V0G703 A0A1A9ZER4 A0A034VHH4 A0A1Q3G3N0 A0A023F3Z6 A0A1Q3G3I4 A0A034VJH9 A0A1Q3G2V5 A0A1B0ATF1 A0A224XAB6 T1HSY1 A0A069DX58 A0A0L0CRM7 A0A0P4VZV7 E0VIP0 Q7YZA4 A0A0Q5ST07 A0A1B6IC32 K7J3N6 Q8IRY0 A0A146MFK9 Q8MT16 A0A146LF02 A0A146LBJ9 A0A1B0CAZ8 B4I9C0 B3P9G0 A0A0Q5SU66
PDB
2J1D
E-value=1.80861e-89,
Score=842
Ontologies
GO
Topology
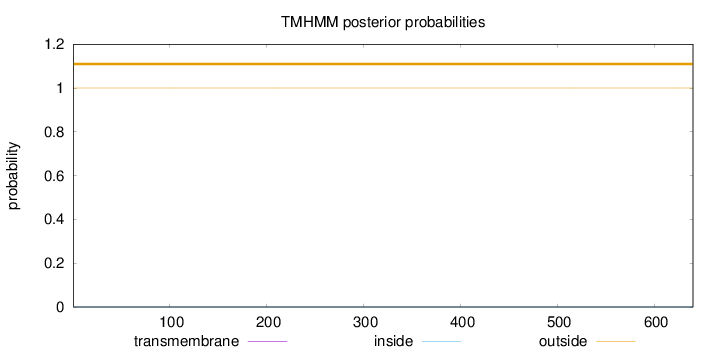
Length:
640
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00205
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00023
outside
1 - 640
Population Genetic Test Statistics
Pi
285.87938
Theta
191.036804
Tajima's D
1.758014
CLR
0.096985
CSRT
0.83970801459927
Interpretation
Uncertain
