Gene
KWMTBOMO10796
Pre Gene Modal
BGIBMGA008436
Annotation
PREDICTED:_disheveled-associated_activator_of_morphogenesis_1_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.101 Mitochondrial Reliability : 1.423
Sequence
CDS
ATGTGCAGCAAGGATCAGATCAATCACATACTCGGGAAGATCAATGTGGGTCTGCCGTCGTGGTCCCGCGGTGTGGAGGCGGTGGAGGCGGCCCGGACGCGTCTGGTGCGTTGGGCATGTGGCGACCCACCTACCGAGCCTGACCCGCCAAGGAAGAAGATGCCGCATGCGCTAAGACGGAGGTGGCCGCTCTGTCCTTGTTTACAGAATGACGAACCGCCCGAGATCACGTACTGCGTGGTGGGCGGCGAAGGAGGTCTCGCCCTCCAAGCTGTCACCCCCACGCATCCCATGCCCCCGGAAGATGAGCTGGACGCCAAGTTTGCGGAGCTGGTCGAGGAGTTGGACCTGACAGCAGTTAACAAGGCAGCGATGATGGCGCTACCGGCTGCCAAGAAATGGCAGATCTACTGCAGTCGCAGGCCGCCACCGGGCCAGGCGCCTCCACTGGCCACCGCCCCACAAGTCGAGGAGTACATCAAGGCCCTTAACGAGATCGCTGATGCATTTGCGTCGTCTGAAGGCGTCCCCCCGACGGAATCCTGCGGCCTACTCGAGGGCCTGAAGACAGCGCTCAGGACGAGGGCTCACAGCTTCGTCCTCCGGTTTATCAAGCAAGGAGGACTCGGTGCCATATTGGACGCGTTGCAGAAGGCGCCCAGGGACGATGCTGTCACGAGACATAACCTTATAGCTGCCATAAAAGCCTTGATGAATAATTCGACTGGTCGTGCTCATGTATTAGCTCATCCCACGAGCATCGACCTGATCGCTCAGTCCTTGGACACGGAGAATGTGAAGACGAAAGTCGCAGCACTTGAGATACTCGGCGCCGTCTGTCTTGTCCCCGGGGGACATAAAAAGGTGCTAGAAGCGATGGTCAACTTCCAAAAGTATTCCGGCGAGAGGGCCCGCTTCCAGAGCATCGTCAACGAGCTGGACCGCAGCACCGGCGCCTACAGGGACGACCTCGGCCTGAAGACGGCCATCATGTCGCTCGTCAACGCCGTGCTCAACTACGGGCCCGGCGAGGAGAGCCTCGAGTTCAGGCTCCATCTGAGATACGAGCTGCTCATGCTGGGGCTTCAACCAGTGATCGAAAAACTCCGTAAATACGAAAACGAGACGTTGGACCGGCACATCGAGTTCTTCGAGATGGTCCGCGGCGAGGACGAGCGCGAGCTGGCGCGGCGCTTCGACCGCGAGCACGTGGACACCAAGAGCGCCTCCGCCATGTTCGAGCTGCTGCGCCGCAAGCTGGGCCACTCCGCCGCCTACCCGCACCTGCTGTCCCTGCTGCAACACCTGCTGCTGCTGCCGCGTGGGTTGTGA
Protein
MCSKDQINHILGKINVGLPSWSRGVEAVEAARTRLVRWACGDPPTEPDPPRKKMPHALRRRWPLCPCLQNDEPPEITYCVVGGEGGLALQAVTPTHPMPPEDELDAKFAELVEELDLTAVNKAAMMALPAAKKWQIYCSRRPPPGQAPPLATAPQVEEYIKALNEIADAFASSEGVPPTESCGLLEGLKTALRTRAHSFVLRFIKQGGLGAILDALQKAPRDDAVTRHNLIAAIKALMNNSTGRAHVLAHPTSIDLIAQSLDTENVKTKVAALEILGAVCLVPGGHKKVLEAMVNFQKYSGERARFQSIVNELDRSTGAYRDDLGLKTAIMSLVNAVLNYGPGEESLEFRLHLRYELLMLGLQPVIEKLRKYENETLDRHIEFFEMVRGEDERELARRFDREHVDTKSASAMFELLRRKLGHSAAYPHLLSLLQHLLLLPRGL
Summary
Uniprot
A0A2A4IZP6
A0A2H1W0G9
A0A0L7KST1
A0A1B6GN34
A0A1B6K981
D2A1M1
+ More
A0A2J7PXF2 A0A1B6H9V9 A0A1Y1L756 A0A1W4WVZ1 A0A1A9VCS2 A0A0L0CRM7 A0A1B0ATF1 A0A182MR58 A0A1A9ZER4 A0A1A9XBL5 A0A1A9WCN2 A0A182X1Y6 A0A182YK93 A0A182LCR6 U5EYQ6 A0A182QN43 A0A182II20 A0A026W786 A0A0N8AGT9 A0A1Y1L9L1 A0A154P3Z0 A0A0J7MUH1 A0A1I8P4R0 A0A158NIU2 A0A1W4WX95 A0A1Q3G3F8 A0A195FDI1 A0A1S4G7X6 A0A1Q3G3N0 A0A1Q3G3I4 A0A1Q3G2V5 T1PIB8 A0A087ZVL2 E2AS93 A0A0T6B243 A0A151J034 A0A336LG97 A0A336MYR9 A0A310SE13 A0A1W4WL92 A0A336N1D8 A0A195CAC0 A0A151WUC1 A0A1W4WX10 B0W4A3 A0A0K8VN44 A0A1I8NCD6 W8ALP6 A0A195BZC4 A0A182UWJ5 W8BAK2 W8AQT8 A0A0P4YH12 A0A0A1XAP4 A0A0P6CY14 A0A0P5B1U4 A0A0P5K2F7 A0A0P5RIW7 A0A034VK79 A0A0P4Y2S6 A0A034VHH4 A0A182UDJ2 A0A0P5IUV4 A0A182P5R7 A0A034VJH9 A0A0P5L7S1 A0A0P5IAC1 A0A0N8CRS2 A0A0C9QEL2 A0A0P5RE29 A0A0P5PEZ3 A0A0P6FXL4 A0A0P5YZA6 A0A0P5XT82 A0A0P5X1X3 E2BJR3 A0A0P5V2W8 A0A0P6DR20 A0A0C9R4G2 A0A0P5V9D8 A0A084VQL4 A0A1B6C417 A0A0C9R184 A0A0P6BGM3 A0A0P5PDQ4 E9GYJ9 A0A0P5ZZQ3 Q16HS4 A0A182GZE7 A0A0M8ZWQ7 A0A0P5XVQ4
A0A2J7PXF2 A0A1B6H9V9 A0A1Y1L756 A0A1W4WVZ1 A0A1A9VCS2 A0A0L0CRM7 A0A1B0ATF1 A0A182MR58 A0A1A9ZER4 A0A1A9XBL5 A0A1A9WCN2 A0A182X1Y6 A0A182YK93 A0A182LCR6 U5EYQ6 A0A182QN43 A0A182II20 A0A026W786 A0A0N8AGT9 A0A1Y1L9L1 A0A154P3Z0 A0A0J7MUH1 A0A1I8P4R0 A0A158NIU2 A0A1W4WX95 A0A1Q3G3F8 A0A195FDI1 A0A1S4G7X6 A0A1Q3G3N0 A0A1Q3G3I4 A0A1Q3G2V5 T1PIB8 A0A087ZVL2 E2AS93 A0A0T6B243 A0A151J034 A0A336LG97 A0A336MYR9 A0A310SE13 A0A1W4WL92 A0A336N1D8 A0A195CAC0 A0A151WUC1 A0A1W4WX10 B0W4A3 A0A0K8VN44 A0A1I8NCD6 W8ALP6 A0A195BZC4 A0A182UWJ5 W8BAK2 W8AQT8 A0A0P4YH12 A0A0A1XAP4 A0A0P6CY14 A0A0P5B1U4 A0A0P5K2F7 A0A0P5RIW7 A0A034VK79 A0A0P4Y2S6 A0A034VHH4 A0A182UDJ2 A0A0P5IUV4 A0A182P5R7 A0A034VJH9 A0A0P5L7S1 A0A0P5IAC1 A0A0N8CRS2 A0A0C9QEL2 A0A0P5RE29 A0A0P5PEZ3 A0A0P6FXL4 A0A0P5YZA6 A0A0P5XT82 A0A0P5X1X3 E2BJR3 A0A0P5V2W8 A0A0P6DR20 A0A0C9R4G2 A0A0P5V9D8 A0A084VQL4 A0A1B6C417 A0A0C9R184 A0A0P6BGM3 A0A0P5PDQ4 E9GYJ9 A0A0P5ZZQ3 Q16HS4 A0A182GZE7 A0A0M8ZWQ7 A0A0P5XVQ4
Pubmed
EMBL
NWSH01004538
PCG64966.1
ODYU01005559
SOQ46527.1
JTDY01006304
KOB66121.1
+ More
GECZ01005917 JAS63852.1 GEBQ01031975 JAT08002.1 KQ971338 EFA01522.1 NEVH01020859 PNF21013.1 GECU01036337 JAS71369.1 GEZM01062702 JAV69522.1 JRES01000104 KNC34099.1 JXJN01003291 AXCM01001179 GANO01001873 JAB57998.1 AXCN02000083 APCN01002159 APCN01002160 KK107432 QOIP01000001 EZA50889.1 RLU26859.1 GDIP01148825 JAJ74577.1 GEZM01062703 GEZM01062701 JAV69521.1 KQ434804 KZC06054.1 LBMM01017307 KMQ84125.1 ADTU01017170 ADTU01017171 ADTU01017172 ADTU01017173 GFDL01000708 JAV34337.1 KQ981677 KYN38104.1 GFDL01000676 JAV34369.1 GFDL01000685 JAV34360.1 GFDL01000898 JAV34147.1 KA647880 AFP62509.1 GL442229 EFN63723.1 LJIG01016132 KRT81519.1 KQ980665 KYN14644.1 UFQS01003390 UFQT01003390 SSX15595.1 SSX34957.1 UFQT01003906 SSX35360.1 KQ766789 OAD53573.1 SSX35359.1 KQ978068 KYM97635.1 KQ982730 KYQ51519.1 DS231836 EDS33188.1 GDHF01018434 GDHF01012012 JAI33880.1 JAI40302.1 GAMC01019688 JAB86867.1 KQ976394 KYM93268.1 GAMC01019686 JAB86869.1 GAMC01019687 JAB86868.1 GDIP01237125 JAI86276.1 GBXI01006539 JAD07753.1 GDIQ01086779 JAN07958.1 GDIP01195314 JAJ28088.1 GDIQ01216281 JAK35444.1 GDIQ01109412 JAL42314.1 GAKP01016984 JAC41968.1 GDIP01237124 JAI86277.1 GAKP01016985 GAKP01016983 JAC41969.1 GDIQ01216280 JAK35445.1 GAKP01016982 JAC41970.1 GDIQ01183194 JAK68531.1 GDIQ01216282 JAK35443.1 GDIP01105271 JAL98443.1 GBYB01001714 JAG71481.1 GDIQ01107221 JAL44505.1 GDIQ01139318 JAL12408.1 GDIQ01041651 GDIQ01001427 JAN53086.1 GDIP01052945 JAM50770.1 GDIP01069587 GDIP01069586 LRGB01001763 JAM34128.1 KZS10665.1 GDIP01078526 JAM25189.1 GL448616 EFN84071.1 GDIP01107822 JAL95892.1 GDIQ01073680 JAN21057.1 GBYB01001716 JAG71483.1 GDIP01118473 JAL85241.1 ATLV01015250 KE525004 KFB40258.1 GEDC01029052 JAS08246.1 GBYB01001720 JAG71487.1 GDIP01016262 JAM87453.1 GDIQ01132276 JAL19450.1 GL732575 EFX75409.1 GDIP01036437 JAM67278.1 CH478145 EAT33802.1 JXUM01099401 KQ564489 KXJ72200.1 KQ435845 KOX71173.1 GDIP01066774 JAM36941.1
GECZ01005917 JAS63852.1 GEBQ01031975 JAT08002.1 KQ971338 EFA01522.1 NEVH01020859 PNF21013.1 GECU01036337 JAS71369.1 GEZM01062702 JAV69522.1 JRES01000104 KNC34099.1 JXJN01003291 AXCM01001179 GANO01001873 JAB57998.1 AXCN02000083 APCN01002159 APCN01002160 KK107432 QOIP01000001 EZA50889.1 RLU26859.1 GDIP01148825 JAJ74577.1 GEZM01062703 GEZM01062701 JAV69521.1 KQ434804 KZC06054.1 LBMM01017307 KMQ84125.1 ADTU01017170 ADTU01017171 ADTU01017172 ADTU01017173 GFDL01000708 JAV34337.1 KQ981677 KYN38104.1 GFDL01000676 JAV34369.1 GFDL01000685 JAV34360.1 GFDL01000898 JAV34147.1 KA647880 AFP62509.1 GL442229 EFN63723.1 LJIG01016132 KRT81519.1 KQ980665 KYN14644.1 UFQS01003390 UFQT01003390 SSX15595.1 SSX34957.1 UFQT01003906 SSX35360.1 KQ766789 OAD53573.1 SSX35359.1 KQ978068 KYM97635.1 KQ982730 KYQ51519.1 DS231836 EDS33188.1 GDHF01018434 GDHF01012012 JAI33880.1 JAI40302.1 GAMC01019688 JAB86867.1 KQ976394 KYM93268.1 GAMC01019686 JAB86869.1 GAMC01019687 JAB86868.1 GDIP01237125 JAI86276.1 GBXI01006539 JAD07753.1 GDIQ01086779 JAN07958.1 GDIP01195314 JAJ28088.1 GDIQ01216281 JAK35444.1 GDIQ01109412 JAL42314.1 GAKP01016984 JAC41968.1 GDIP01237124 JAI86277.1 GAKP01016985 GAKP01016983 JAC41969.1 GDIQ01216280 JAK35445.1 GAKP01016982 JAC41970.1 GDIQ01183194 JAK68531.1 GDIQ01216282 JAK35443.1 GDIP01105271 JAL98443.1 GBYB01001714 JAG71481.1 GDIQ01107221 JAL44505.1 GDIQ01139318 JAL12408.1 GDIQ01041651 GDIQ01001427 JAN53086.1 GDIP01052945 JAM50770.1 GDIP01069587 GDIP01069586 LRGB01001763 JAM34128.1 KZS10665.1 GDIP01078526 JAM25189.1 GL448616 EFN84071.1 GDIP01107822 JAL95892.1 GDIQ01073680 JAN21057.1 GBYB01001716 JAG71483.1 GDIP01118473 JAL85241.1 ATLV01015250 KE525004 KFB40258.1 GEDC01029052 JAS08246.1 GBYB01001720 JAG71487.1 GDIP01016262 JAM87453.1 GDIQ01132276 JAL19450.1 GL732575 EFX75409.1 GDIP01036437 JAM67278.1 CH478145 EAT33802.1 JXUM01099401 KQ564489 KXJ72200.1 KQ435845 KOX71173.1 GDIP01066774 JAM36941.1
Proteomes
UP000218220
UP000037510
UP000007266
UP000235965
UP000192223
UP000078200
+ More
UP000037069 UP000092460 UP000075883 UP000092445 UP000092443 UP000091820 UP000076407 UP000076408 UP000075882 UP000075886 UP000075840 UP000053097 UP000279307 UP000076502 UP000036403 UP000095300 UP000005205 UP000078541 UP000005203 UP000000311 UP000078492 UP000078542 UP000075809 UP000002320 UP000095301 UP000078540 UP000075903 UP000075902 UP000075885 UP000076858 UP000008237 UP000030765 UP000000305 UP000008820 UP000069940 UP000249989 UP000053105
UP000037069 UP000092460 UP000075883 UP000092445 UP000092443 UP000091820 UP000076407 UP000076408 UP000075882 UP000075886 UP000075840 UP000053097 UP000279307 UP000076502 UP000036403 UP000095300 UP000005205 UP000078541 UP000005203 UP000000311 UP000078492 UP000078542 UP000075809 UP000002320 UP000095301 UP000078540 UP000075903 UP000075902 UP000075885 UP000076858 UP000008237 UP000030765 UP000000305 UP000008820 UP000069940 UP000249989 UP000053105
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2A4IZP6
A0A2H1W0G9
A0A0L7KST1
A0A1B6GN34
A0A1B6K981
D2A1M1
+ More
A0A2J7PXF2 A0A1B6H9V9 A0A1Y1L756 A0A1W4WVZ1 A0A1A9VCS2 A0A0L0CRM7 A0A1B0ATF1 A0A182MR58 A0A1A9ZER4 A0A1A9XBL5 A0A1A9WCN2 A0A182X1Y6 A0A182YK93 A0A182LCR6 U5EYQ6 A0A182QN43 A0A182II20 A0A026W786 A0A0N8AGT9 A0A1Y1L9L1 A0A154P3Z0 A0A0J7MUH1 A0A1I8P4R0 A0A158NIU2 A0A1W4WX95 A0A1Q3G3F8 A0A195FDI1 A0A1S4G7X6 A0A1Q3G3N0 A0A1Q3G3I4 A0A1Q3G2V5 T1PIB8 A0A087ZVL2 E2AS93 A0A0T6B243 A0A151J034 A0A336LG97 A0A336MYR9 A0A310SE13 A0A1W4WL92 A0A336N1D8 A0A195CAC0 A0A151WUC1 A0A1W4WX10 B0W4A3 A0A0K8VN44 A0A1I8NCD6 W8ALP6 A0A195BZC4 A0A182UWJ5 W8BAK2 W8AQT8 A0A0P4YH12 A0A0A1XAP4 A0A0P6CY14 A0A0P5B1U4 A0A0P5K2F7 A0A0P5RIW7 A0A034VK79 A0A0P4Y2S6 A0A034VHH4 A0A182UDJ2 A0A0P5IUV4 A0A182P5R7 A0A034VJH9 A0A0P5L7S1 A0A0P5IAC1 A0A0N8CRS2 A0A0C9QEL2 A0A0P5RE29 A0A0P5PEZ3 A0A0P6FXL4 A0A0P5YZA6 A0A0P5XT82 A0A0P5X1X3 E2BJR3 A0A0P5V2W8 A0A0P6DR20 A0A0C9R4G2 A0A0P5V9D8 A0A084VQL4 A0A1B6C417 A0A0C9R184 A0A0P6BGM3 A0A0P5PDQ4 E9GYJ9 A0A0P5ZZQ3 Q16HS4 A0A182GZE7 A0A0M8ZWQ7 A0A0P5XVQ4
A0A2J7PXF2 A0A1B6H9V9 A0A1Y1L756 A0A1W4WVZ1 A0A1A9VCS2 A0A0L0CRM7 A0A1B0ATF1 A0A182MR58 A0A1A9ZER4 A0A1A9XBL5 A0A1A9WCN2 A0A182X1Y6 A0A182YK93 A0A182LCR6 U5EYQ6 A0A182QN43 A0A182II20 A0A026W786 A0A0N8AGT9 A0A1Y1L9L1 A0A154P3Z0 A0A0J7MUH1 A0A1I8P4R0 A0A158NIU2 A0A1W4WX95 A0A1Q3G3F8 A0A195FDI1 A0A1S4G7X6 A0A1Q3G3N0 A0A1Q3G3I4 A0A1Q3G2V5 T1PIB8 A0A087ZVL2 E2AS93 A0A0T6B243 A0A151J034 A0A336LG97 A0A336MYR9 A0A310SE13 A0A1W4WL92 A0A336N1D8 A0A195CAC0 A0A151WUC1 A0A1W4WX10 B0W4A3 A0A0K8VN44 A0A1I8NCD6 W8ALP6 A0A195BZC4 A0A182UWJ5 W8BAK2 W8AQT8 A0A0P4YH12 A0A0A1XAP4 A0A0P6CY14 A0A0P5B1U4 A0A0P5K2F7 A0A0P5RIW7 A0A034VK79 A0A0P4Y2S6 A0A034VHH4 A0A182UDJ2 A0A0P5IUV4 A0A182P5R7 A0A034VJH9 A0A0P5L7S1 A0A0P5IAC1 A0A0N8CRS2 A0A0C9QEL2 A0A0P5RE29 A0A0P5PEZ3 A0A0P6FXL4 A0A0P5YZA6 A0A0P5XT82 A0A0P5X1X3 E2BJR3 A0A0P5V2W8 A0A0P6DR20 A0A0C9R4G2 A0A0P5V9D8 A0A084VQL4 A0A1B6C417 A0A0C9R184 A0A0P6BGM3 A0A0P5PDQ4 E9GYJ9 A0A0P5ZZQ3 Q16HS4 A0A182GZE7 A0A0M8ZWQ7 A0A0P5XVQ4
PDB
4YDH
E-value=2.56422e-23,
Score=270
Ontologies
GO
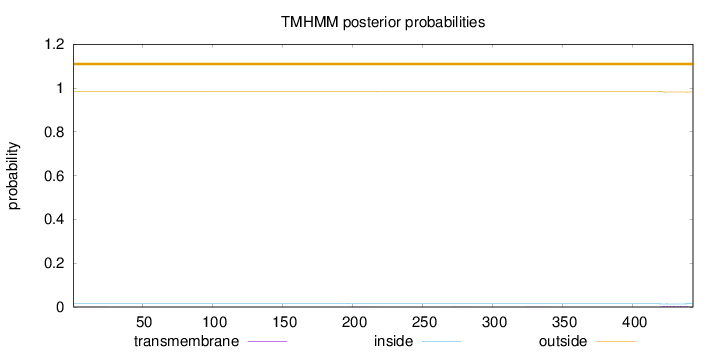
Topology
Length:
443
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.10284
Exp number, first 60 AAs:
0.00245
Total prob of N-in:
0.01537
outside
1 - 443
Population Genetic Test Statistics
Pi
186.492331
Theta
155.15688
Tajima's D
0.583118
CLR
0.702328
CSRT
0.538523073846308
Interpretation
Uncertain
