Pre Gene Modal
BGIBMGA008387
Annotation
Cleavage_and_polyadenylation_specificity_factor_[Operophtera_brumata]
Full name
Integrator complex subunit 11
Location in the cell
Cytoplasmic Reliability : 2.671
Sequence
CDS
ATGCCGGATATTAAGATTACGCCACTCGGAGCGGGCCAGGATGTTGGTCGGAGCTGCATCTTGCTAACCATGGGTGGCAAAAACATAATGTTAGATTGTGGTATGCATATGGGCTACAACGATGAAAGAAGATTTCCTGATTTCTCTTACATTGTCCCGGAAGGCCCTATAACGAGTCAGATTGACTGCGTTATTATATCACATTTTCACCTAGATCACTGCGGTGCACTGCCCTATATGTCCGAAATGATTGGTTATACCGGCCCTATTTACATGACACACCCGACTAAAGCCATAGCACCAATCCTTCTCGAGGATATGAGGAAAGTCGCAGTGGAGCGCAAAGGTGAGTCAAACTTCTTCACTTCCCAAATGATTAAAGATTGTATGAAAAAAGTGACAGCTGTGACTCTCCATCAATCAGTAATGGTGGATAATGAATTAGAAATCAAAGCTTATTACGCAGGACATGTCTTAGGAGCTGCAATGTTCTGGATCAGAGTAGGGTCACAATCTGTGGTTTACACAGGTGATTACAACATGACACCTGATAGGCACTTAGGAGCCGCCTGGATTGACAAATGTAGACCTGATCTGCTCATAACAGAATCCACATATGCAACAACCATAAGAGATTCAAAACGCTGTCGTGAAAGAGATTTTCTCAAAAAAGTTCATGAATGTGTAGAAAAAGGAGGTAAGGTTCTGATTCCGGTGTTTGCATTAGGAAGGGCACAGGAACTTTGTATTTTATTGGAAACATATTGGGATCGTATGAATCTTAAATATCCAGTTTACTTTGCTCTTGGACTGACAGAAAAAGCAAACAATTACTATAAAATGTTTATAACATGGACCAATCAAAAAATAAGAAAGACATTTGTTCAAAGAAATATGTTTGACTTTAAGCATATTAAACCATTTGACAAGACATACATTGATAATCCCGGTGCTATGGTAGTTTTTGCTACGCCTGGAATGTTGCATGCTGGATTGTCACTAAACATCTTTAAGAAATGGGCTCCTTATGAGCAGAACATGGTGATTATGCCAGGATTCTGTGTGCAAGGCACAGTCGGACACAAGATCCTCAATGGAGCAAAGAAGGTTGAATTTGAAAACAGACAAGTGGTTGAAGTTAAAATGGCAGTAGAATACATGTCATTTTCAGCTCATGCTGATGCAAAAGGAATTATGCAATTGATACAATATTGTGAACCCAAAAATGTATTGTTGGTACATGGTGAAGCTCAAAAAATGGAGTTTTTGAAAGATAAAATAGAGAAGGAATTCAAAATTAGTTGTTTTATGCCAGCTAATGGTGAAACATGTGTTATAAATACTCCAACAACCATTCCTATTGATGTCTCGTTGAGGTTGCTGAAAGCTGAAGCAGTGAGATACAATGCCCAACCACCTGATCCAAAACGCAGGCGAGTTGTTAATGGGATTCTGTGTGTGAAAGATAACAGGCTATCATTACTAGACATTGATGAGATGTGTGATGAAATCGGAATAAACAGGCATATCATTAGATTCACAAGTACAGTTCGTTTTGAGGATCCGGGGCCAGCTATCAAGACTGCTGAAAAATTGCAGAATTTACTGACGGAAAAATTAGAAGGCTGGACTATAACTATATCGGAAGGAAACATATCAGTTGAGTCAGTACTCATCAAGGTAGAAGGTGAAGATGATAGCACAAAAAACATTTATGTTTCTTGGACAAACCAAGACGAAGACTTAGGTAGCTATATATTAGGGCTCCTGCAATCAATGGTTCAATATAGTCCAATGAAAGAATAA
Protein
MPDIKITPLGAGQDVGRSCILLTMGGKNIMLDCGMHMGYNDERRFPDFSYIVPEGPITSQIDCVIISHFHLDHCGALPYMSEMIGYTGPIYMTHPTKAIAPILLEDMRKVAVERKGESNFFTSQMIKDCMKKVTAVTLHQSVMVDNELEIKAYYAGHVLGAAMFWIRVGSQSVVYTGDYNMTPDRHLGAAWIDKCRPDLLITESTYATTIRDSKRCRERDFLKKVHECVEKGGKVLIPVFALGRAQELCILLETYWDRMNLKYPVYFALGLTEKANNYYKMFITWTNQKIRKTFVQRNMFDFKHIKPFDKTYIDNPGAMVVFATPGMLHAGLSLNIFKKWAPYEQNMVIMPGFCVQGTVGHKILNGAKKVEFENRQVVEVKMAVEYMSFSAHADAKGIMQLIQYCEPKNVLLVHGEAQKMEFLKDKIEKEFKISCFMPANGETCVINTPTTIPIDVSLRLLKAEAVRYNAQPPDPKRRRVVNGILCVKDNRLSLLDIDEMCDEIGINRHIIRFTSTVRFEDPGPAIKTAEKLQNLLTEKLEGWTITISEGNISVESVLIKVEGEDDSTKNIYVSWTNQDEDLGSYILGLLQSMVQYSPMKE
Summary
Description
Catalytic component of the Integrator complex, a complex involved in the transcription of small nuclear RNAs (snRNA) and their 3'-box-dependent processing (By similarity). Mediates the snRNAs 3' cleavage (By similarity). Involved in the 3'-end processing of the U7 snRNA, and also the spliceosomal snRNAs U1, U2, U4 and U5 (PubMed:21078872, PubMed:23097424). May mediate recruitment of cytoplasmic dynein to the nuclear envelope, probably as component of the INT complex (By similarity).
Subunit
Belongs to the multiprotein complex Integrator, at least composed of IntS1, IntS2, IntS3, IntS4, omd/IntS5, IntS6, defl/IntS7, IntS8, IntS9, IntS10, IntS11, IntS12, asun/IntS13 and IntS14. Within the complex, interacts with IntS12.
Similarity
Belongs to the metallo-beta-lactamase superfamily. RNA-metabolizing metallo-beta-lactamase-like family. INTS11 subfamily.
Keywords
Complete proteome
Cytoplasm
Hydrolase
Nucleus
Reference proteome
Feature
chain Integrator complex subunit 11
Uniprot
H9JFU0
A0A2A4JJC9
A0A2H1VFQ1
A0A0L7LBD0
A0A212FC57
S4P7Y7
+ More
A0A194PCB6 A0A0N1IP68 A0A1B0CS58 A0A1L8DH16 A0A1Y1KLE6 A0A067QZP5 A0A2P8YMQ0 Q17BY3 A0A2M4AJE4 B0WXD2 D6WSY0 A0A240PLI8 N6TD56 A0A182NAT9 A0A195FIZ8 A0A182PG30 W5JL07 A0A195E4S5 A0A195AW54 A0A1Q3F3F8 A0A158P276 A0A182W8Z3 A0A182LVZ7 A0A182K1F6 E2AU94 A0A151X234 A0A182QZ49 A0A0J7KSS2 A0A182RR95 A0A154P487 A0A182YMQ9 A0A026WZY5 A0A182KYX1 Q7Q184 A0A182HRG3 A0A182TES4 A0A1W4UPW8 B4PPK8 B3MTI3 B4R083 A0A151IQZ9 B4HZE1 Q9VAH9 B3P5D8 T1PIJ9 A0A1W4X7R4 A0A2A3EP12 B4M5T1 A0A0L0BMX6 A0A336MK34 W8BWY4 A0A1A9UQ76 Q29AN7 A0A1I8PXZ8 A0A232FAT6 A0A1A9W6J6 A0A182F1N9 K7IUU0 B4NBL1 A0A0K8U3C4 A0A088AL35 E2B377 B4G664 A0A0N1ITA5 A0A0C9REI0 A0A1W7R5M0 B4K612 A0A0M3QYR2 A0A1A9XMV4 A0A146KSZ9 A0A310SC00 A0A3B0JPS9 B4JYM6 A0A182V783 A0A182XLT8 A0A2R7WHK4 A0A0V0GBB5 T1HGA7 A0A023F3V0 A0A1B6MGT3 A0A0K8TKD2 A0A1B6D3P6 A0A182H2C0 A0A224XLA7 A0A1S4EK63 A0A182STI6 A0A034WIR9 E0VNH8 A0A0A9YW13 A0A2M4BJV1 A0A2M4BJ27 B7PHB3 A0A131Y1L2 A0A210QTM6
A0A194PCB6 A0A0N1IP68 A0A1B0CS58 A0A1L8DH16 A0A1Y1KLE6 A0A067QZP5 A0A2P8YMQ0 Q17BY3 A0A2M4AJE4 B0WXD2 D6WSY0 A0A240PLI8 N6TD56 A0A182NAT9 A0A195FIZ8 A0A182PG30 W5JL07 A0A195E4S5 A0A195AW54 A0A1Q3F3F8 A0A158P276 A0A182W8Z3 A0A182LVZ7 A0A182K1F6 E2AU94 A0A151X234 A0A182QZ49 A0A0J7KSS2 A0A182RR95 A0A154P487 A0A182YMQ9 A0A026WZY5 A0A182KYX1 Q7Q184 A0A182HRG3 A0A182TES4 A0A1W4UPW8 B4PPK8 B3MTI3 B4R083 A0A151IQZ9 B4HZE1 Q9VAH9 B3P5D8 T1PIJ9 A0A1W4X7R4 A0A2A3EP12 B4M5T1 A0A0L0BMX6 A0A336MK34 W8BWY4 A0A1A9UQ76 Q29AN7 A0A1I8PXZ8 A0A232FAT6 A0A1A9W6J6 A0A182F1N9 K7IUU0 B4NBL1 A0A0K8U3C4 A0A088AL35 E2B377 B4G664 A0A0N1ITA5 A0A0C9REI0 A0A1W7R5M0 B4K612 A0A0M3QYR2 A0A1A9XMV4 A0A146KSZ9 A0A310SC00 A0A3B0JPS9 B4JYM6 A0A182V783 A0A182XLT8 A0A2R7WHK4 A0A0V0GBB5 T1HGA7 A0A023F3V0 A0A1B6MGT3 A0A0K8TKD2 A0A1B6D3P6 A0A182H2C0 A0A224XLA7 A0A1S4EK63 A0A182STI6 A0A034WIR9 E0VNH8 A0A0A9YW13 A0A2M4BJV1 A0A2M4BJ27 B7PHB3 A0A131Y1L2 A0A210QTM6
EC Number
3.1.27.-
Pubmed
19121390
26227816
22118469
23622113
26354079
28004739
+ More
24845553 29403074 17510324 18362917 19820115 23537049 20920257 23761445 21347285 20798317 25244985 24508170 20966253 12364791 14747013 17210077 17994087 17550304 10731132 12537572 12537569 21078872 23097424 25315136 26108605 24495485 15632085 28648823 20075255 26823975 25474469 26369729 26483478 25348373 20566863 25401762 28812685
24845553 29403074 17510324 18362917 19820115 23537049 20920257 23761445 21347285 20798317 25244985 24508170 20966253 12364791 14747013 17210077 17994087 17550304 10731132 12537572 12537569 21078872 23097424 25315136 26108605 24495485 15632085 28648823 20075255 26823975 25474469 26369729 26483478 25348373 20566863 25401762 28812685
EMBL
BABH01001612
NWSH01001369
PCG71513.1
ODYU01002318
SOQ39659.1
JTDY01001817
+ More
KOB72797.1 AGBW02009218 OWR51326.1 GAIX01004384 JAA88176.1 KQ459606 KPI90921.1 KQ460556 KPJ14009.1 AJWK01025650 AJWK01025651 GFDF01008341 JAV05743.1 GEZM01086240 JAV59657.1 KK852854 KDR15001.1 PYGN01000487 PSN45526.1 CH477314 EAT43837.1 GGFK01007584 MBW40905.1 DS232162 EDS36462.1 KQ971352 EFA06334.1 APGK01035338 KB740923 KB632333 ENN78279.1 ERL92707.1 KQ981523 KYN40346.1 ADMH02001134 ETN63983.1 KQ979657 KYN19907.1 KQ976731 KYM76194.1 GFDL01012978 JAV22067.1 ADTU01007008 AXCM01000135 GL442815 EFN62951.1 KQ982580 KYQ54467.1 AXCN02000659 LBMM01003507 KMQ93447.1 KQ434809 KZC06667.1 KK107054 EZA61587.1 AAAB01008980 EAA13845.4 APCN01001701 CM000160 EDW98258.1 CH902623 EDV30573.1 CM000364 EDX14889.1 KQ976755 KYN08598.1 CH480819 EDW53398.1 AE014297 AY061097 AAL28645.1 CH954182 EDV53188.1 KA648479 AFP63108.1 KZ288209 PBC32992.1 CH940652 EDW59007.1 JRES01001623 KNC21362.1 UFQS01001164 UFQT01000573 UFQT01001164 SSX09301.1 SSX25388.1 SSX29203.1 GAMC01002798 JAC03758.1 CM000070 EAL27312.1 NNAY01000550 OXU27732.1 AAZX01002635 CH964232 EDW81175.1 GDHF01031107 JAI21207.1 GL445305 EFN89859.1 CH479179 EDW23817.1 KQ435843 KOX71282.1 GBYB01015019 JAG84786.1 GEHC01001203 JAV46442.1 CH933806 EDW16249.1 CP012526 ALC48059.1 GDHC01019018 JAP99610.1 KQ764051 OAD54615.1 OUUW01000007 SPP82913.1 CH916377 EDV90788.1 KK854825 PTY19048.1 GECL01000899 JAP05225.1 ACPB03015950 GBBI01002631 JAC16081.1 GEBQ01004817 JAT35160.1 GDAI01002879 JAI14724.1 GEDC01022799 GEDC01016998 JAS14499.1 JAS20300.1 JXUM01023801 KQ560671 KXJ81343.1 GFTR01007216 JAW09210.1 GAKP01005289 JAC53663.1 DS235339 EEB14934.1 GBHO01006417 JAG37187.1 GGFJ01003927 MBW53068.1 GGFJ01003928 MBW53069.1 ABJB010582038 DS711851 EEC05985.1 GEFM01003880 JAP71916.1 NEDP02001979 OWF52067.1
KOB72797.1 AGBW02009218 OWR51326.1 GAIX01004384 JAA88176.1 KQ459606 KPI90921.1 KQ460556 KPJ14009.1 AJWK01025650 AJWK01025651 GFDF01008341 JAV05743.1 GEZM01086240 JAV59657.1 KK852854 KDR15001.1 PYGN01000487 PSN45526.1 CH477314 EAT43837.1 GGFK01007584 MBW40905.1 DS232162 EDS36462.1 KQ971352 EFA06334.1 APGK01035338 KB740923 KB632333 ENN78279.1 ERL92707.1 KQ981523 KYN40346.1 ADMH02001134 ETN63983.1 KQ979657 KYN19907.1 KQ976731 KYM76194.1 GFDL01012978 JAV22067.1 ADTU01007008 AXCM01000135 GL442815 EFN62951.1 KQ982580 KYQ54467.1 AXCN02000659 LBMM01003507 KMQ93447.1 KQ434809 KZC06667.1 KK107054 EZA61587.1 AAAB01008980 EAA13845.4 APCN01001701 CM000160 EDW98258.1 CH902623 EDV30573.1 CM000364 EDX14889.1 KQ976755 KYN08598.1 CH480819 EDW53398.1 AE014297 AY061097 AAL28645.1 CH954182 EDV53188.1 KA648479 AFP63108.1 KZ288209 PBC32992.1 CH940652 EDW59007.1 JRES01001623 KNC21362.1 UFQS01001164 UFQT01000573 UFQT01001164 SSX09301.1 SSX25388.1 SSX29203.1 GAMC01002798 JAC03758.1 CM000070 EAL27312.1 NNAY01000550 OXU27732.1 AAZX01002635 CH964232 EDW81175.1 GDHF01031107 JAI21207.1 GL445305 EFN89859.1 CH479179 EDW23817.1 KQ435843 KOX71282.1 GBYB01015019 JAG84786.1 GEHC01001203 JAV46442.1 CH933806 EDW16249.1 CP012526 ALC48059.1 GDHC01019018 JAP99610.1 KQ764051 OAD54615.1 OUUW01000007 SPP82913.1 CH916377 EDV90788.1 KK854825 PTY19048.1 GECL01000899 JAP05225.1 ACPB03015950 GBBI01002631 JAC16081.1 GEBQ01004817 JAT35160.1 GDAI01002879 JAI14724.1 GEDC01022799 GEDC01016998 JAS14499.1 JAS20300.1 JXUM01023801 KQ560671 KXJ81343.1 GFTR01007216 JAW09210.1 GAKP01005289 JAC53663.1 DS235339 EEB14934.1 GBHO01006417 JAG37187.1 GGFJ01003927 MBW53068.1 GGFJ01003928 MBW53069.1 ABJB010582038 DS711851 EEC05985.1 GEFM01003880 JAP71916.1 NEDP02001979 OWF52067.1
Proteomes
UP000005204
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
+ More
UP000092461 UP000027135 UP000245037 UP000008820 UP000002320 UP000007266 UP000075880 UP000019118 UP000030742 UP000075884 UP000078541 UP000075885 UP000000673 UP000078492 UP000078540 UP000005205 UP000075920 UP000075883 UP000075881 UP000000311 UP000075809 UP000075886 UP000036403 UP000075900 UP000076502 UP000076408 UP000053097 UP000075882 UP000007062 UP000075840 UP000075902 UP000192221 UP000002282 UP000007801 UP000000304 UP000078542 UP000001292 UP000000803 UP000008711 UP000095301 UP000192223 UP000242457 UP000008792 UP000037069 UP000078200 UP000001819 UP000095300 UP000215335 UP000091820 UP000069272 UP000002358 UP000007798 UP000005203 UP000008237 UP000008744 UP000053105 UP000009192 UP000092553 UP000092443 UP000268350 UP000001070 UP000075903 UP000076407 UP000015103 UP000069940 UP000249989 UP000079169 UP000075901 UP000009046 UP000001555 UP000242188
UP000092461 UP000027135 UP000245037 UP000008820 UP000002320 UP000007266 UP000075880 UP000019118 UP000030742 UP000075884 UP000078541 UP000075885 UP000000673 UP000078492 UP000078540 UP000005205 UP000075920 UP000075883 UP000075881 UP000000311 UP000075809 UP000075886 UP000036403 UP000075900 UP000076502 UP000076408 UP000053097 UP000075882 UP000007062 UP000075840 UP000075902 UP000192221 UP000002282 UP000007801 UP000000304 UP000078542 UP000001292 UP000000803 UP000008711 UP000095301 UP000192223 UP000242457 UP000008792 UP000037069 UP000078200 UP000001819 UP000095300 UP000215335 UP000091820 UP000069272 UP000002358 UP000007798 UP000005203 UP000008237 UP000008744 UP000053105 UP000009192 UP000092553 UP000092443 UP000268350 UP000001070 UP000075903 UP000076407 UP000015103 UP000069940 UP000249989 UP000079169 UP000075901 UP000009046 UP000001555 UP000242188
Pfam
Interpro
IPR001279
Metallo-B-lactamas
+ More
IPR041897 INTS11-like_MBL-fold
IPR022712 Beta_Casp
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR011108 RMMBL
IPR003210 Signal_recog_particle_SRP14
IPR010678 Digest_organ_expansion_fac-prd
IPR009018 Signal_recog_particle_SRP9/14
IPR001589 Actinin_actin-bd_CS
IPR036452 Ribo_hydro-like
IPR001910 Inosine/uridine_hydrolase_dom
IPR001494 Importin-beta_N
IPR021133 HEAT_type_2
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR040122 Importin_beta
IPR041897 INTS11-like_MBL-fold
IPR022712 Beta_Casp
IPR036866 RibonucZ/Hydroxyglut_hydro
IPR011108 RMMBL
IPR003210 Signal_recog_particle_SRP14
IPR010678 Digest_organ_expansion_fac-prd
IPR009018 Signal_recog_particle_SRP9/14
IPR001589 Actinin_actin-bd_CS
IPR036452 Ribo_hydro-like
IPR001910 Inosine/uridine_hydrolase_dom
IPR001494 Importin-beta_N
IPR021133 HEAT_type_2
IPR016024 ARM-type_fold
IPR011989 ARM-like
IPR040122 Importin_beta
Gene 3D
ProteinModelPortal
H9JFU0
A0A2A4JJC9
A0A2H1VFQ1
A0A0L7LBD0
A0A212FC57
S4P7Y7
+ More
A0A194PCB6 A0A0N1IP68 A0A1B0CS58 A0A1L8DH16 A0A1Y1KLE6 A0A067QZP5 A0A2P8YMQ0 Q17BY3 A0A2M4AJE4 B0WXD2 D6WSY0 A0A240PLI8 N6TD56 A0A182NAT9 A0A195FIZ8 A0A182PG30 W5JL07 A0A195E4S5 A0A195AW54 A0A1Q3F3F8 A0A158P276 A0A182W8Z3 A0A182LVZ7 A0A182K1F6 E2AU94 A0A151X234 A0A182QZ49 A0A0J7KSS2 A0A182RR95 A0A154P487 A0A182YMQ9 A0A026WZY5 A0A182KYX1 Q7Q184 A0A182HRG3 A0A182TES4 A0A1W4UPW8 B4PPK8 B3MTI3 B4R083 A0A151IQZ9 B4HZE1 Q9VAH9 B3P5D8 T1PIJ9 A0A1W4X7R4 A0A2A3EP12 B4M5T1 A0A0L0BMX6 A0A336MK34 W8BWY4 A0A1A9UQ76 Q29AN7 A0A1I8PXZ8 A0A232FAT6 A0A1A9W6J6 A0A182F1N9 K7IUU0 B4NBL1 A0A0K8U3C4 A0A088AL35 E2B377 B4G664 A0A0N1ITA5 A0A0C9REI0 A0A1W7R5M0 B4K612 A0A0M3QYR2 A0A1A9XMV4 A0A146KSZ9 A0A310SC00 A0A3B0JPS9 B4JYM6 A0A182V783 A0A182XLT8 A0A2R7WHK4 A0A0V0GBB5 T1HGA7 A0A023F3V0 A0A1B6MGT3 A0A0K8TKD2 A0A1B6D3P6 A0A182H2C0 A0A224XLA7 A0A1S4EK63 A0A182STI6 A0A034WIR9 E0VNH8 A0A0A9YW13 A0A2M4BJV1 A0A2M4BJ27 B7PHB3 A0A131Y1L2 A0A210QTM6
A0A194PCB6 A0A0N1IP68 A0A1B0CS58 A0A1L8DH16 A0A1Y1KLE6 A0A067QZP5 A0A2P8YMQ0 Q17BY3 A0A2M4AJE4 B0WXD2 D6WSY0 A0A240PLI8 N6TD56 A0A182NAT9 A0A195FIZ8 A0A182PG30 W5JL07 A0A195E4S5 A0A195AW54 A0A1Q3F3F8 A0A158P276 A0A182W8Z3 A0A182LVZ7 A0A182K1F6 E2AU94 A0A151X234 A0A182QZ49 A0A0J7KSS2 A0A182RR95 A0A154P487 A0A182YMQ9 A0A026WZY5 A0A182KYX1 Q7Q184 A0A182HRG3 A0A182TES4 A0A1W4UPW8 B4PPK8 B3MTI3 B4R083 A0A151IQZ9 B4HZE1 Q9VAH9 B3P5D8 T1PIJ9 A0A1W4X7R4 A0A2A3EP12 B4M5T1 A0A0L0BMX6 A0A336MK34 W8BWY4 A0A1A9UQ76 Q29AN7 A0A1I8PXZ8 A0A232FAT6 A0A1A9W6J6 A0A182F1N9 K7IUU0 B4NBL1 A0A0K8U3C4 A0A088AL35 E2B377 B4G664 A0A0N1ITA5 A0A0C9REI0 A0A1W7R5M0 B4K612 A0A0M3QYR2 A0A1A9XMV4 A0A146KSZ9 A0A310SC00 A0A3B0JPS9 B4JYM6 A0A182V783 A0A182XLT8 A0A2R7WHK4 A0A0V0GBB5 T1HGA7 A0A023F3V0 A0A1B6MGT3 A0A0K8TKD2 A0A1B6D3P6 A0A182H2C0 A0A224XLA7 A0A1S4EK63 A0A182STI6 A0A034WIR9 E0VNH8 A0A0A9YW13 A0A2M4BJV1 A0A2M4BJ27 B7PHB3 A0A131Y1L2 A0A210QTM6
PDB
2I7V
E-value=7.96634e-94,
Score=879
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Cytoplasm
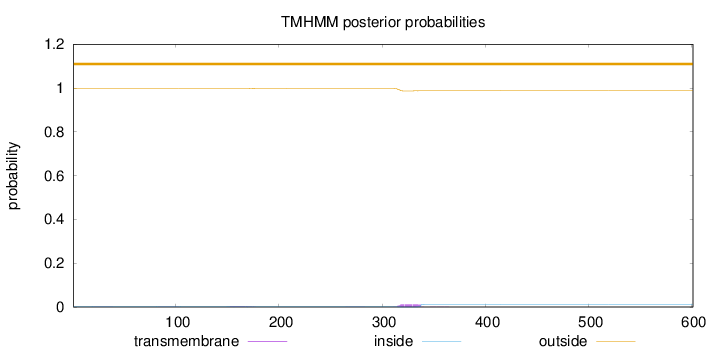
Length:
601
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.35016
Exp number, first 60 AAs:
0.00883
Total prob of N-in:
0.00330
outside
1 - 601
Population Genetic Test Statistics
Pi
281.325673
Theta
214.06771
Tajima's D
1.012663
CLR
0.003245
CSRT
0.660416979151042
Interpretation
Uncertain
