Gene
KWMTBOMO10790
Pre Gene Modal
BGIBMGA008432
Annotation
PREDICTED:_tubulin_polyglutamylase_TTLL6_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.275 Mitochondrial Reliability : 1.023
Sequence
CDS
ATGGCCGTGCTAATGAGGAGGATGATAAACTCGGAAAATATAAACGATAAATACGTAGAGCGCGCCATCAAGAAACGTTATTGGGTTTATTCTGCTCAGAATGACGTCAACAATCCTGACGGTTTGTTAAAGCACGTGCATCGCGTCCTAAAACGAATCGGTTACGAAAAAACCACGAACGAATCCGATTGGAACTTGCTATGGTCCCACGATTATCCTTTTCGAGTACTGTACCCTAATTTACACAGTTTAAAACCAAATCAGAAAGTCAACCACTACCCCGGAGCGGGTTTCGTTACGAACAAAGTAGACCTTGCCACGACTGAATCGAAATACATCCCGAAGGCGTTCAAACTGCCACAGAATAAAGAAGATTTTTTAAAATACGCCGCTGAGAACGAAGGCGCCTTGTTTTTGGAAAAGCACAATCAACACAGAGGTGTTTACTTAAAAAACGTGAGCGAAATAAATCTGGCTGGCGGGGAAAGTTTCGTACAAGAATACGTGCAAAGACCGTTCTTAGTCGACGGACACAAGTTTGATATTGGCGTGTACGTAGTTCTAACCTCGGTTAACCCCTTGAGAGCATATTGGTACAAAGGAGATGTCCTCTTCAGGTACTGTCCGGCAAAGTATTATCCTTTCGATCCAAAGGATTTGGATAAATATGTCATTAGGGACGACTACCTCCCTACCTGGGAGGTACCTTCGCTGTCGCATCCTTACACTGCCCTTGGGTTTAGCATGAGAGATGCTTTCGACATTTACGCTAAAACAAAGGGGAAAGACCCTACACAGATGTGGAGGGAAATACAAAACGCTATAGCTGAGTTATTTGTGAAAAAAGAGCATCATATTGTCGAAACTTTGAAATCGTATCCGTCAACAGAAAACTTCTTCGAACTAATGCGTTTTGATCTGGTCGTCGATGAAGATCTAAAAGTGTATTTGTTAGAAGCCAACATGTCGCCCAACTTGTCGTCTGCTCACTACCCTCCCAACCAGCTGTTGTACGAGCAGGTTCTGTACAACTTGTTCGGACTGGTTGGCATAGCTGATCATATCCAGGAGAATAGTCTTGGAGATTCAGCTGCTCAGAACATGGTGTCTTCACAGAAAAACATAGCCGTATGGGGCGAAATATGCACGACGACTTGTAGAGATAATTGTCACATGAATCTAATATGTGAACTCTGTCGTCCCTGTCTGAGCGCAAGGTTGCGAAGCGTCTTCCTTAAGGCCCATAAGGAGTTTCTTCATAAGGGAGACTTTAGGAGGTTGTTCCCACCGCCTATGAAAGCCAATGCTACTTTAAAAGAAGAGGACTTGAAGCACTTGAACAAACCAAACAGACTTCAGTACTTATGGTACCAGGGAAAATGCAATGTGGATATCACTTGGTGTACATAA
Protein
MAVLMRRMINSENINDKYVERAIKKRYWVYSAQNDVNNPDGLLKHVHRVLKRIGYEKTTNESDWNLLWSHDYPFRVLYPNLHSLKPNQKVNHYPGAGFVTNKVDLATTESKYIPKAFKLPQNKEDFLKYAAENEGALFLEKHNQHRGVYLKNVSEINLAGGESFVQEYVQRPFLVDGHKFDIGVYVVLTSVNPLRAYWYKGDVLFRYCPAKYYPFDPKDLDKYVIRDDYLPTWEVPSLSHPYTALGFSMRDAFDIYAKTKGKDPTQMWREIQNAIAELFVKKEHHIVETLKSYPSTENFFELMRFDLVVDEDLKVYLLEANMSPNLSSAHYPPNQLLYEQVLYNLFGLVGIADHIQENSLGDSAAQNMVSSQKNIAVWGEICTTTCRDNCHMNLICELCRPCLSARLRSVFLKAHKEFLHKGDFRRLFPPPMKANATLKEEDLKHLNKPNRLQYLWYQGKCNVDITWCT
Summary
Uniprot
H9JFY5
A0A3S2P685
A0A212FC66
A0A0N1I7L8
A0A2H1VFN9
A0A2A4JIW4
+ More
A0A194PCJ8 A0A154PHL4 K7IQF8 A0A232EXE1 D6WP53 A0A0J7KMT0 A0A151X5I6 A0A195DIB9 A0A2A3EBJ2 A0A158NRI8 A0A195BE45 A0A195FU15 E2BD39 A0A0L7QM28 N6U974 A0A026W7A3 U4UR99 F4WBM3 W5JWA9 A0A182Y903 A0A182QX78 A0A084WPB6 B0WNF8 A0A182MM81 B3M295 A0A2J7Q368 A0A182GQF0 A0A182J0Q7 A0A182UE80 A0A2J7Q374 Q17H42 A0A182NCV4 A0A182V661 A0A182XF72 A0A1S4F2V9 A0NDN6 A0A182PLH6 A0A336L223 A0A182RI20 A0A087ZT70 A0A182FAF8 A0A182KZS5 A0A182HVP3 A0A182JQ72 A0A1W4VVN6 A0A0M9A4F4 A0A1B0CSE5 A0A1W4VWD6 A0A0M4F7A0 Q9VGW2 A0A067R1T6 A0A1I8MP31 D3TNX0 B3P1J6 A0A0L7LBM6 A0A0L0BNE0 A0A1A9ZK50 B4PLD8 A0A1B0BPA5 A0A1A9XSQ2 B4JHM0 C3XS25 B4GDZ8 A0A1A9UX29 A0A182W389 B4NJY7 A0A0P4WA57 A0A1S3J7I1 A0A1S3I8R5 A0A3B0K5C1 A0A1J1IMD4 B4HIP6 R7VCQ4 B4KC94 C3XS26 A0A151I766 A0A1D2NHW0 E9J8N3 W8B330 E9G6D5 A0A0P6EXC0 A0A0P5PYW8 A0A0N8BT97 A0A1I8PNT0 A0A0P6CFP9 A0A0A9X8G8 A0A2S2N7P3 A0A164UR34 B4M5I6 V4CKY4 A0A0P6HCP9 A0A1B0DDN9 A0A0P5C3Z2 A0A0P4YDF3
A0A194PCJ8 A0A154PHL4 K7IQF8 A0A232EXE1 D6WP53 A0A0J7KMT0 A0A151X5I6 A0A195DIB9 A0A2A3EBJ2 A0A158NRI8 A0A195BE45 A0A195FU15 E2BD39 A0A0L7QM28 N6U974 A0A026W7A3 U4UR99 F4WBM3 W5JWA9 A0A182Y903 A0A182QX78 A0A084WPB6 B0WNF8 A0A182MM81 B3M295 A0A2J7Q368 A0A182GQF0 A0A182J0Q7 A0A182UE80 A0A2J7Q374 Q17H42 A0A182NCV4 A0A182V661 A0A182XF72 A0A1S4F2V9 A0NDN6 A0A182PLH6 A0A336L223 A0A182RI20 A0A087ZT70 A0A182FAF8 A0A182KZS5 A0A182HVP3 A0A182JQ72 A0A1W4VVN6 A0A0M9A4F4 A0A1B0CSE5 A0A1W4VWD6 A0A0M4F7A0 Q9VGW2 A0A067R1T6 A0A1I8MP31 D3TNX0 B3P1J6 A0A0L7LBM6 A0A0L0BNE0 A0A1A9ZK50 B4PLD8 A0A1B0BPA5 A0A1A9XSQ2 B4JHM0 C3XS25 B4GDZ8 A0A1A9UX29 A0A182W389 B4NJY7 A0A0P4WA57 A0A1S3J7I1 A0A1S3I8R5 A0A3B0K5C1 A0A1J1IMD4 B4HIP6 R7VCQ4 B4KC94 C3XS26 A0A151I766 A0A1D2NHW0 E9J8N3 W8B330 E9G6D5 A0A0P6EXC0 A0A0P5PYW8 A0A0N8BT97 A0A1I8PNT0 A0A0P6CFP9 A0A0A9X8G8 A0A2S2N7P3 A0A164UR34 B4M5I6 V4CKY4 A0A0P6HCP9 A0A1B0DDN9 A0A0P5C3Z2 A0A0P4YDF3
Pubmed
19121390
22118469
26354079
20075255
28648823
18362917
+ More
19820115 21347285 20798317 23537049 24508170 30249741 21719571 20920257 23761445 25244985 24438588 17994087 26483478 17510324 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 25315136 20353571 26227816 26108605 17550304 18563158 26383154 23254933 27289101 21282665 24495485 21292972 25401762
19820115 21347285 20798317 23537049 24508170 30249741 21719571 20920257 23761445 25244985 24438588 17994087 26483478 17510324 12364791 14747013 17210077 20966253 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 25315136 20353571 26227816 26108605 17550304 18563158 26383154 23254933 27289101 21282665 24495485 21292972 25401762
EMBL
BABH01001609
BABH01001610
BABH01001611
BABH01001612
RSAL01000298
RVE42799.1
+ More
AGBW02009218 OWR51325.1 KQ460556 KPJ14008.1 ODYU01002318 SOQ39658.1 NWSH01001369 PCG71514.1 KQ459606 KPI90922.1 KQ434905 KZC11297.1 NNAY01001737 OXU23076.1 KQ971352 EFA07282.1 LBMM01005228 KMQ91688.1 KQ982498 KYQ55675.1 KQ980824 KYN12633.1 KZ288292 PBC29088.1 ADTU01024048 KQ976511 KYM82440.1 KQ981280 KYN43379.1 GL447563 EFN86381.1 KQ414894 KOC59698.1 APGK01035293 KB740923 ENN78250.1 KK107364 QOIP01000007 EZA51935.1 RLU20371.1 KB632333 ERL92681.1 GL888066 EGI68301.1 ADMH02000180 ETN67465.1 AXCN02001207 ATLV01025016 KE525367 KFB52060.1 DS232010 EDS31612.1 AXCM01001830 CH902617 EDV42286.1 NEVH01019069 PNF23035.1 JXUM01080395 KQ563183 KXJ74316.1 PNF23038.1 CH477253 EAT45971.1 AAAB01008898 EAU76857.3 UFQS01001681 UFQT01001681 SSX11840.1 SSX31404.1 APCN01005809 KQ435736 KOX77017.1 AJWK01025954 CP012526 ALC47707.1 AE014297 AY061104 AAF54561.1 AAL28652.1 KK853091 KDR11546.1 CCAG010019253 EZ423122 ADD19398.1 CH954181 EDV49595.1 JTDY01001817 KOB72800.1 JRES01001605 KNC21547.1 CM000160 EDW96845.1 JXJN01017876 CH916369 EDV93859.1 GG666456 EEN69493.1 CH479182 EDW33833.1 CH964272 EDW83989.1 GDRN01059911 JAI65461.1 OUUW01000007 SPP83210.1 CVRI01000054 CRL00246.1 CH480815 EDW42693.1 AMQN01004320 KB293181 ELU16337.1 CH933806 EDW13703.2 EEN69494.1 KQ978430 KYM94012.1 LJIJ01000035 ODN04837.1 GL769036 EFZ10825.1 GAMC01015119 GAMC01015117 JAB91438.1 GL732533 EFX84915.1 GDIQ01069215 JAN25522.1 GDIQ01121528 JAL30198.1 GDIQ01147083 JAL04643.1 GDIP01094852 GDIP01015972 JAM87743.1 GBHO01027320 JAG16284.1 GGMR01000479 MBY13098.1 LRGB01001579 KZS11596.1 CH940652 EDW58912.2 KB200129 ESP02910.1 GDIQ01021336 JAN73401.1 AJVK01032116 GDIP01176450 GDIP01094853 JAJ46952.1 GDIP01231790 JAI91611.1
AGBW02009218 OWR51325.1 KQ460556 KPJ14008.1 ODYU01002318 SOQ39658.1 NWSH01001369 PCG71514.1 KQ459606 KPI90922.1 KQ434905 KZC11297.1 NNAY01001737 OXU23076.1 KQ971352 EFA07282.1 LBMM01005228 KMQ91688.1 KQ982498 KYQ55675.1 KQ980824 KYN12633.1 KZ288292 PBC29088.1 ADTU01024048 KQ976511 KYM82440.1 KQ981280 KYN43379.1 GL447563 EFN86381.1 KQ414894 KOC59698.1 APGK01035293 KB740923 ENN78250.1 KK107364 QOIP01000007 EZA51935.1 RLU20371.1 KB632333 ERL92681.1 GL888066 EGI68301.1 ADMH02000180 ETN67465.1 AXCN02001207 ATLV01025016 KE525367 KFB52060.1 DS232010 EDS31612.1 AXCM01001830 CH902617 EDV42286.1 NEVH01019069 PNF23035.1 JXUM01080395 KQ563183 KXJ74316.1 PNF23038.1 CH477253 EAT45971.1 AAAB01008898 EAU76857.3 UFQS01001681 UFQT01001681 SSX11840.1 SSX31404.1 APCN01005809 KQ435736 KOX77017.1 AJWK01025954 CP012526 ALC47707.1 AE014297 AY061104 AAF54561.1 AAL28652.1 KK853091 KDR11546.1 CCAG010019253 EZ423122 ADD19398.1 CH954181 EDV49595.1 JTDY01001817 KOB72800.1 JRES01001605 KNC21547.1 CM000160 EDW96845.1 JXJN01017876 CH916369 EDV93859.1 GG666456 EEN69493.1 CH479182 EDW33833.1 CH964272 EDW83989.1 GDRN01059911 JAI65461.1 OUUW01000007 SPP83210.1 CVRI01000054 CRL00246.1 CH480815 EDW42693.1 AMQN01004320 KB293181 ELU16337.1 CH933806 EDW13703.2 EEN69494.1 KQ978430 KYM94012.1 LJIJ01000035 ODN04837.1 GL769036 EFZ10825.1 GAMC01015119 GAMC01015117 JAB91438.1 GL732533 EFX84915.1 GDIQ01069215 JAN25522.1 GDIQ01121528 JAL30198.1 GDIQ01147083 JAL04643.1 GDIP01094852 GDIP01015972 JAM87743.1 GBHO01027320 JAG16284.1 GGMR01000479 MBY13098.1 LRGB01001579 KZS11596.1 CH940652 EDW58912.2 KB200129 ESP02910.1 GDIQ01021336 JAN73401.1 AJVK01032116 GDIP01176450 GDIP01094853 JAJ46952.1 GDIP01231790 JAI91611.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000218220
UP000053268
+ More
UP000076502 UP000002358 UP000215335 UP000007266 UP000036403 UP000075809 UP000078492 UP000242457 UP000005205 UP000078540 UP000078541 UP000008237 UP000053825 UP000019118 UP000053097 UP000279307 UP000030742 UP000007755 UP000000673 UP000076408 UP000075886 UP000030765 UP000002320 UP000075883 UP000007801 UP000235965 UP000069940 UP000249989 UP000075880 UP000075902 UP000008820 UP000075884 UP000075903 UP000076407 UP000007062 UP000075885 UP000075900 UP000005203 UP000069272 UP000075882 UP000075840 UP000075881 UP000192221 UP000053105 UP000092461 UP000092553 UP000000803 UP000027135 UP000095301 UP000092444 UP000008711 UP000037510 UP000037069 UP000092445 UP000002282 UP000092460 UP000092443 UP000001070 UP000001554 UP000008744 UP000078200 UP000075920 UP000007798 UP000085678 UP000268350 UP000183832 UP000001292 UP000014760 UP000009192 UP000078542 UP000094527 UP000000305 UP000095300 UP000076858 UP000008792 UP000030746 UP000092462
UP000076502 UP000002358 UP000215335 UP000007266 UP000036403 UP000075809 UP000078492 UP000242457 UP000005205 UP000078540 UP000078541 UP000008237 UP000053825 UP000019118 UP000053097 UP000279307 UP000030742 UP000007755 UP000000673 UP000076408 UP000075886 UP000030765 UP000002320 UP000075883 UP000007801 UP000235965 UP000069940 UP000249989 UP000075880 UP000075902 UP000008820 UP000075884 UP000075903 UP000076407 UP000007062 UP000075885 UP000075900 UP000005203 UP000069272 UP000075882 UP000075840 UP000075881 UP000192221 UP000053105 UP000092461 UP000092553 UP000000803 UP000027135 UP000095301 UP000092444 UP000008711 UP000037510 UP000037069 UP000092445 UP000002282 UP000092460 UP000092443 UP000001070 UP000001554 UP000008744 UP000078200 UP000075920 UP000007798 UP000085678 UP000268350 UP000183832 UP000001292 UP000014760 UP000009192 UP000078542 UP000094527 UP000000305 UP000095300 UP000076858 UP000008792 UP000030746 UP000092462
Interpro
SUPFAM
SSF54928
SSF54928
Gene 3D
ProteinModelPortal
H9JFY5
A0A3S2P685
A0A212FC66
A0A0N1I7L8
A0A2H1VFN9
A0A2A4JIW4
+ More
A0A194PCJ8 A0A154PHL4 K7IQF8 A0A232EXE1 D6WP53 A0A0J7KMT0 A0A151X5I6 A0A195DIB9 A0A2A3EBJ2 A0A158NRI8 A0A195BE45 A0A195FU15 E2BD39 A0A0L7QM28 N6U974 A0A026W7A3 U4UR99 F4WBM3 W5JWA9 A0A182Y903 A0A182QX78 A0A084WPB6 B0WNF8 A0A182MM81 B3M295 A0A2J7Q368 A0A182GQF0 A0A182J0Q7 A0A182UE80 A0A2J7Q374 Q17H42 A0A182NCV4 A0A182V661 A0A182XF72 A0A1S4F2V9 A0NDN6 A0A182PLH6 A0A336L223 A0A182RI20 A0A087ZT70 A0A182FAF8 A0A182KZS5 A0A182HVP3 A0A182JQ72 A0A1W4VVN6 A0A0M9A4F4 A0A1B0CSE5 A0A1W4VWD6 A0A0M4F7A0 Q9VGW2 A0A067R1T6 A0A1I8MP31 D3TNX0 B3P1J6 A0A0L7LBM6 A0A0L0BNE0 A0A1A9ZK50 B4PLD8 A0A1B0BPA5 A0A1A9XSQ2 B4JHM0 C3XS25 B4GDZ8 A0A1A9UX29 A0A182W389 B4NJY7 A0A0P4WA57 A0A1S3J7I1 A0A1S3I8R5 A0A3B0K5C1 A0A1J1IMD4 B4HIP6 R7VCQ4 B4KC94 C3XS26 A0A151I766 A0A1D2NHW0 E9J8N3 W8B330 E9G6D5 A0A0P6EXC0 A0A0P5PYW8 A0A0N8BT97 A0A1I8PNT0 A0A0P6CFP9 A0A0A9X8G8 A0A2S2N7P3 A0A164UR34 B4M5I6 V4CKY4 A0A0P6HCP9 A0A1B0DDN9 A0A0P5C3Z2 A0A0P4YDF3
A0A194PCJ8 A0A154PHL4 K7IQF8 A0A232EXE1 D6WP53 A0A0J7KMT0 A0A151X5I6 A0A195DIB9 A0A2A3EBJ2 A0A158NRI8 A0A195BE45 A0A195FU15 E2BD39 A0A0L7QM28 N6U974 A0A026W7A3 U4UR99 F4WBM3 W5JWA9 A0A182Y903 A0A182QX78 A0A084WPB6 B0WNF8 A0A182MM81 B3M295 A0A2J7Q368 A0A182GQF0 A0A182J0Q7 A0A182UE80 A0A2J7Q374 Q17H42 A0A182NCV4 A0A182V661 A0A182XF72 A0A1S4F2V9 A0NDN6 A0A182PLH6 A0A336L223 A0A182RI20 A0A087ZT70 A0A182FAF8 A0A182KZS5 A0A182HVP3 A0A182JQ72 A0A1W4VVN6 A0A0M9A4F4 A0A1B0CSE5 A0A1W4VWD6 A0A0M4F7A0 Q9VGW2 A0A067R1T6 A0A1I8MP31 D3TNX0 B3P1J6 A0A0L7LBM6 A0A0L0BNE0 A0A1A9ZK50 B4PLD8 A0A1B0BPA5 A0A1A9XSQ2 B4JHM0 C3XS25 B4GDZ8 A0A1A9UX29 A0A182W389 B4NJY7 A0A0P4WA57 A0A1S3J7I1 A0A1S3I8R5 A0A3B0K5C1 A0A1J1IMD4 B4HIP6 R7VCQ4 B4KC94 C3XS26 A0A151I766 A0A1D2NHW0 E9J8N3 W8B330 E9G6D5 A0A0P6EXC0 A0A0P5PYW8 A0A0N8BT97 A0A1I8PNT0 A0A0P6CFP9 A0A0A9X8G8 A0A2S2N7P3 A0A164UR34 B4M5I6 V4CKY4 A0A0P6HCP9 A0A1B0DDN9 A0A0P5C3Z2 A0A0P4YDF3
PDB
4YLS
E-value=4.9137e-20,
Score=241
Ontologies
GO
Topology
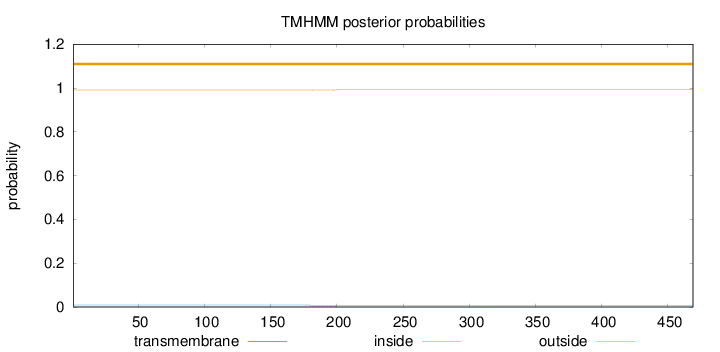
Length:
469
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0386300000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00886
outside
1 - 469
Population Genetic Test Statistics
Pi
208.394604
Theta
167.588672
Tajima's D
0.552403
CLR
32.96708
CSRT
0.527073646317684
Interpretation
Uncertain
