Gene
KWMTBOMO10789
Pre Gene Modal
BGIBMGA008431
Annotation
PREDICTED:_inhibin_beta_chain?_partial_[Bombyx_mori]
Full name
Inhibin beta chain
Alternative Name
Activin beta chain
Location in the cell
Extracellular Reliability : 1.194 Mitochondrial Reliability : 1.088 Nuclear Reliability : 1.453
Sequence
CDS
ATGAAAGAGGATGATCACAACTCACAACGGAGACAATCACTGGAAAGAGCAACAGAACTTCAGAACAACAGAACAGCAATCAGTCCGACTCTGAACGGACAAAGATTGATCGAGTTCCGGCTCGGCCAGGAGGGCGACTCGAATCGAGGGGAGGGACTGGAGGTCGCTTCCGCCGAACTCATGGTGCGCGCCGAGACGGTGCCCCATGCGCGGCACCTGCGCTATACGCTGTGGGCCTTCACCGTCGGAGACAACGCGACCGTCGGCGCGCTCGCCGGCGCCACGAGGAGGGACGCGGCGCGCGCTTGGCAGCGGCTCGATGTGACCGCGGCTGCGCAGCGCTGGGCCGCCGCCGGGGCGCTGACGCCGCTGCGTCTGCTGTTGGACTGCAGCGGGTGCGGCGGCCGCGTGCGCCTGCGGCTGGGCGGCGGACAGGCGGCGCGGCCCTTGCTGCGTCTTCGCCTGATGCCGCCGCCCGCTCGTCGTCGACGGGCGCTGGACTGCGACGGGTCGGAGCATGGCCGGTGTTGTCGACAGACCTACTACGTGAGCTTCCGCGCGCTCGGCTGGGACGACTGGATCGTCGCGCCAGAGGGCTACTACGCCAACTACTGTCGTGGGGCCTGCGCGCCCTTCCTCAACTACCACTCGCAAGTGGTGGAGGCGGCGCGGCTCGAGCGCGCCGCGTGCTGTGCGCCTGTGCGCTTTTCGGCGCTGTCGCTCATCTACTTCGGCGCCGACTCGAACATCATCAAACGTGACCTGCCCGAGATGGTGGTCGAGGAGTGCGACTGCCCGTAG
Protein
MKEDDHNSQRRQSLERATELQNNRTAISPTLNGQRLIEFRLGQEGDSNRGEGLEVASAELMVRAETVPHARHLRYTLWAFTVGDNATVGALAGATRRDAARAWQRLDVTAAAQRWAAAGALTPLRLLLDCSGCGGRVRLRLGGGQAARPLLRLRLMPPPARRRRALDCDGSEHGRCCRQTYYVSFRALGWDDWIVAPEGYYANYCRGACAPFLNYHSQVVEAARLERAACCAPVRFSALSLIYFGADSNIIKRDLPEMVVEECDCP
Summary
Description
Controls several aspects of neuronal morphogenesis; essential for optic lobe development, EcR-B1 expression in larval brains, mushroom body remodeling, dorsal neuron morphogenesis and motoneuron axon guidance. Ligands Actbeta and daw act redundantly through the Activin receptor Babo and its transcriptional mediator Smad2 (Smox), to regulate neuroblast numbers and proliferation rates in the developing larval brain.
Subunit
Homodimer or heterodimer; disulfide-linked.
Similarity
Belongs to the TGF-beta family.
Keywords
Cleavage on pair of basic residues
Complete proteome
Developmental protein
Disulfide bond
Glycoprotein
Growth factor
Reference proteome
Secreted
Signal
Feature
chain Inhibin beta chain
Uniprot
H9JFY4
A0A212FC65
A0A0N1PIX3
A0A0L7LBD4
A0A1B0GID9
A0A067RFS6
+ More
A0A1S3D6N1 A0A2J7PJR1 A0A2P8Z9S3 A0A2R7WM44 D2A422 U4UA25 K7IVN3 E2C3J0 A0A158P356 A0A195CDA4 A0A195EZL3 A0A026WXS5 A0A195DN26 T1I4N3 A0A3L8E2R8 A0A2S2PKN5 A0A2H8TV86 J9KCT9 A0A2S2Q675 A0A154PJ56 A0A0J9S1D6 A0A0J9S166 A0A087ZS78 A0A310SST3 A0A0Q9XFD8 N6TEJ7 A0A0Q9XRU3 A0A0R1EC83 B4PW45 O61643 A0A1W4V580 B3P9P6 B4L7C6 B4IKR2 A0A0N8NZU7 A0A0P9BLR6 A0A3B0K6M1 B4MEZ5 B4JZN5 D0Z798 A0A151WYV9 Q29CT1 A0A2R5LC94 T1JDG1
A0A1S3D6N1 A0A2J7PJR1 A0A2P8Z9S3 A0A2R7WM44 D2A422 U4UA25 K7IVN3 E2C3J0 A0A158P356 A0A195CDA4 A0A195EZL3 A0A026WXS5 A0A195DN26 T1I4N3 A0A3L8E2R8 A0A2S2PKN5 A0A2H8TV86 J9KCT9 A0A2S2Q675 A0A154PJ56 A0A0J9S1D6 A0A0J9S166 A0A087ZS78 A0A310SST3 A0A0Q9XFD8 N6TEJ7 A0A0Q9XRU3 A0A0R1EC83 B4PW45 O61643 A0A1W4V580 B3P9P6 B4L7C6 B4IKR2 A0A0N8NZU7 A0A0P9BLR6 A0A3B0K6M1 B4MEZ5 B4JZN5 D0Z798 A0A151WYV9 Q29CT1 A0A2R5LC94 T1JDG1
Pubmed
EMBL
BABH01001606
AGBW02009218
OWR51324.1
KQ460556
KPJ14007.1
JTDY01001817
+ More
KOB72798.1 AJWK01013757 KK852498 KDR22706.1 NEVH01024946 PNF16575.1 PYGN01000134 PSN53245.1 KK854977 PTY20070.1 KQ971348 EFA05602.2 KB631865 ERL86800.1 GL452320 EFN77528.1 ADTU01007753 KQ977935 KYM98690.1 KQ981905 KYN33566.1 KK107087 EZA59949.1 KQ980713 KYN14278.1 ACPB03011117 ACPB03011118 QOIP01000001 RLU26753.1 GGMR01017380 MBY29999.1 GFXV01005273 MBW17078.1 ABLF02028224 ABLF02028225 ABLF02028228 GGMS01004016 MBY73219.1 KQ434931 KZC11843.1 CM002916 KMZ07378.1 KMZ07377.1 KQ760382 OAD60698.1 CH933813 KRG07280.1 APGK01041006 APGK01041007 KB740986 ENN76158.1 KRG07281.1 CM000161 KRK04993.1 EDW99350.1 KRK04990.1 KRK04991.1 KRK04992.1 AE014135 AY121686 AF454392 AF054822 CH954184 EDV45209.1 EDW10920.2 KRG07282.1 KRG07283.1 CH480855 EDW52652.1 CH902645 KPU75316.1 CH902665 KPU72656.1 OUUW01000017 SPP88933.1 CH940665 EDW71096.2 KRF85342.1 KRF85343.1 KRF85344.1 CH916380 EDV90901.1 CM000831 ACY70530.1 KQ982649 KYQ53082.1 CH475412 EAL29247.2 KRT05385.1 GGLE01003018 MBY07144.1 JH432105
KOB72798.1 AJWK01013757 KK852498 KDR22706.1 NEVH01024946 PNF16575.1 PYGN01000134 PSN53245.1 KK854977 PTY20070.1 KQ971348 EFA05602.2 KB631865 ERL86800.1 GL452320 EFN77528.1 ADTU01007753 KQ977935 KYM98690.1 KQ981905 KYN33566.1 KK107087 EZA59949.1 KQ980713 KYN14278.1 ACPB03011117 ACPB03011118 QOIP01000001 RLU26753.1 GGMR01017380 MBY29999.1 GFXV01005273 MBW17078.1 ABLF02028224 ABLF02028225 ABLF02028228 GGMS01004016 MBY73219.1 KQ434931 KZC11843.1 CM002916 KMZ07378.1 KMZ07377.1 KQ760382 OAD60698.1 CH933813 KRG07280.1 APGK01041006 APGK01041007 KB740986 ENN76158.1 KRG07281.1 CM000161 KRK04993.1 EDW99350.1 KRK04990.1 KRK04991.1 KRK04992.1 AE014135 AY121686 AF454392 AF054822 CH954184 EDV45209.1 EDW10920.2 KRG07282.1 KRG07283.1 CH480855 EDW52652.1 CH902645 KPU75316.1 CH902665 KPU72656.1 OUUW01000017 SPP88933.1 CH940665 EDW71096.2 KRF85342.1 KRF85343.1 KRF85344.1 CH916380 EDV90901.1 CM000831 ACY70530.1 KQ982649 KYQ53082.1 CH475412 EAL29247.2 KRT05385.1 GGLE01003018 MBY07144.1 JH432105
Proteomes
UP000005204
UP000007151
UP000053240
UP000037510
UP000092461
UP000027135
+ More
UP000079169 UP000235965 UP000245037 UP000007266 UP000030742 UP000002358 UP000008237 UP000005205 UP000078542 UP000078541 UP000053097 UP000078492 UP000015103 UP000279307 UP000007819 UP000076502 UP000005203 UP000009192 UP000019118 UP000002282 UP000000803 UP000192221 UP000008711 UP000001292 UP000007801 UP000268350 UP000008792 UP000001070 UP000075809 UP000001819
UP000079169 UP000235965 UP000245037 UP000007266 UP000030742 UP000002358 UP000008237 UP000005205 UP000078542 UP000078541 UP000053097 UP000078492 UP000015103 UP000279307 UP000007819 UP000076502 UP000005203 UP000009192 UP000019118 UP000002282 UP000000803 UP000192221 UP000008711 UP000001292 UP000007801 UP000268350 UP000008792 UP000001070 UP000075809 UP000001819
Interpro
SUPFAM
SSF57501
SSF57501
Gene 3D
ProteinModelPortal
H9JFY4
A0A212FC65
A0A0N1PIX3
A0A0L7LBD4
A0A1B0GID9
A0A067RFS6
+ More
A0A1S3D6N1 A0A2J7PJR1 A0A2P8Z9S3 A0A2R7WM44 D2A422 U4UA25 K7IVN3 E2C3J0 A0A158P356 A0A195CDA4 A0A195EZL3 A0A026WXS5 A0A195DN26 T1I4N3 A0A3L8E2R8 A0A2S2PKN5 A0A2H8TV86 J9KCT9 A0A2S2Q675 A0A154PJ56 A0A0J9S1D6 A0A0J9S166 A0A087ZS78 A0A310SST3 A0A0Q9XFD8 N6TEJ7 A0A0Q9XRU3 A0A0R1EC83 B4PW45 O61643 A0A1W4V580 B3P9P6 B4L7C6 B4IKR2 A0A0N8NZU7 A0A0P9BLR6 A0A3B0K6M1 B4MEZ5 B4JZN5 D0Z798 A0A151WYV9 Q29CT1 A0A2R5LC94 T1JDG1
A0A1S3D6N1 A0A2J7PJR1 A0A2P8Z9S3 A0A2R7WM44 D2A422 U4UA25 K7IVN3 E2C3J0 A0A158P356 A0A195CDA4 A0A195EZL3 A0A026WXS5 A0A195DN26 T1I4N3 A0A3L8E2R8 A0A2S2PKN5 A0A2H8TV86 J9KCT9 A0A2S2Q675 A0A154PJ56 A0A0J9S1D6 A0A0J9S166 A0A087ZS78 A0A310SST3 A0A0Q9XFD8 N6TEJ7 A0A0Q9XRU3 A0A0R1EC83 B4PW45 O61643 A0A1W4V580 B3P9P6 B4L7C6 B4IKR2 A0A0N8NZU7 A0A0P9BLR6 A0A3B0K6M1 B4MEZ5 B4JZN5 D0Z798 A0A151WYV9 Q29CT1 A0A2R5LC94 T1JDG1
PDB
5HLY
E-value=1.20933e-17,
Score=218
Ontologies
GO
PANTHER
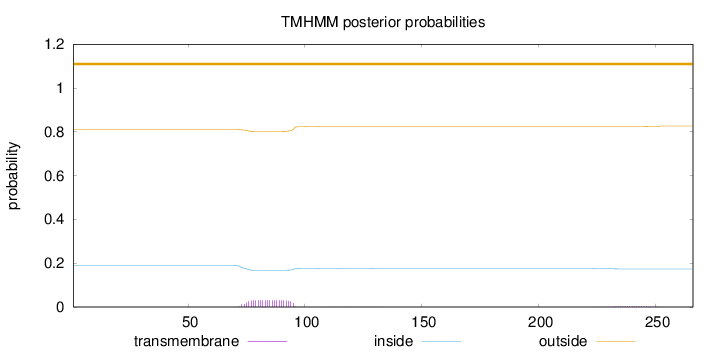
Topology
Subcellular location
Secreted
Length:
266
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.70911
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.18926
outside
1 - 266
Population Genetic Test Statistics
Pi
221.43362
Theta
180.532052
Tajima's D
0.635876
CLR
0.229444
CSRT
0.557122143892805
Interpretation
Uncertain
