Gene
KWMTBOMO10785
Pre Gene Modal
BGIBMGA008389
Annotation
Inhibitor_of_nuclear_factor_kappa_B_kinase_beta_subunit_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 3.367
Sequence
CDS
ATGAACGATATAGTGTTCGTTGGCGATTGGATTAAAGACAGAGTACTGGGATCAGGCAGCTTTGGGACAGTAGTGCTGTGGAAACATAAAAAGACCGAAGAGAAACTCGCCATCAAAACTTGTAAATGGGGCGATGAACTCACTGACAAACACAGAGACAGGTGGACGAAGGAGGTTGAAATGTTGCAAAGCTGCCGAAATCCAAACATCGTTGGATCGAAACCCCTTCCATCTGAATTTATAAAGGGCTTAGAGCGAGGCAATCCTTCAGGTCTACCTATAATGTGTATGGAATACTGCAGCGGAGGTGATCTTAGGCAGCAATTGAATAAACCCGAATCATGTTCTGGACTGCGAGAAATACAAGTTCGTCAGATTCTCAGCGATATCAGAAATGCAATGAAGTTCTTACATCAAAAGAAAATAACTCACCGTGATCTAAAACCAGAAAATATTGTTATGAATATTGTAGAGAATACAGAGCAAAACAATATTTCTCCTCAAACTGAACCAAGAGTCATATACAAACTGATAGATCTAGGTTATGCGAAGGAGATTGACTTTAATTCCGTGTGTGCCAGCTTTGTTGGTACTCTTCAATATTTAGCACCAGAGCTACTGTACAGTAAAACATATAGCAATTCTGTTGACTTTTGGTCATTTGGTTTGCTTGCTTTTGAAATAATTTGTGGGAAAAGGCCTTTCCTTCCATTCATGTCTCCTGCACAGTGGATGCCTCACGTTGAAAAGAAAACACATGAAAATATTTGTGTTTATGAAACTTTTCATGGAGATATTGAATATTCAAACAATATATTTCCTGAAAATCATATGTCAGCACCTTTTAAAGTATTAATGGAAGATTGGCTTAGAATAGCTCTGGAATGGGATCCTAAACTGAGAGGCAGAGATTCTCATACAAGAGTCAGATTTAATATTCCTACTGAGCAAAGAGAAGTAAGCAGTAACATCATCATTTATGACCTACTGGAAAATATTTTATCTCGGAAGATAATAAAAATATTTTCTGTACCTACTCTGTCACGACTAGCATACCAAATCAATGATACTACAATGGTTTCTGATTTGAAGAAATGGATTGGGCAAGATACAAGTATGGATATAGCAGACTTAATCTTGATATGTCCAAATACTTTTACAGAACTTGGAGAAATGGACATTGTGTCAAACTATTGGAGAGAACACTGTAACACAATGGTATACTTGTTCAATAAAACATACATTCTTCAGAAGAACATAGAACCAGAAGTGCCAAAATCAGTACAAAGATGCTTGGAGCATCCTAAAGCTTTGTACAATCACAAAAATAGCCAAACTTTATATAAGAATAGCTTGTATTTTGTAAAGAATCAATCTGAAATTTATGAATCACTCATTAATGGGTTACTTGTTAGAGGCGAATCACTAAAAAATGAGGGCAAACAATTGCTACTAAAACATAATGCAGTTGATAAAAGTATAAGTAATCTTCTAGCAAAGGAAGAAATAATGAAAAGGCTGATTGAAACTGGTAAGGAGCAAGTAAACAAATTAAGAGACAGTGGAAAGGGCATAAACATGCTTGGAGGTTTCAACAAAACATTCCAAGACACAGATGAGCATTTCCTAAAAATCAGTAAACTCCAACAAGCCTGGAATCAATTGTCTGTTAGACTCACATCTGCTAAACGACGTTGTAATGAAATTGTGTCAGATGATATAAAAAACTTTGTAGCCAAATATAATTACCAAGCAGTTTATGCCAATGCATTCAAAATATACATTATGGCAAGGAAAAGTGAAATTATCGGAAAAGAAAAACACTGTGATGACATTATGAAAATTTGCTACAACTGTCTCAAATTGAGAAGTAAAATATTGCTTGAAATTCATCACCAGCCCTTTGTTGTGAAGCTGATGGATTTGGGTACTGAATTTTCCAAGATATCCGACATTGTGACCAAGGCTGCAGAAAACACAGAAAAATTAACCAAAGACCTCAGTGATTACATTGATGACCTGAATAATTGTATATGGTCCACCATAAACTTGGCAGTTAGTGATGGGGATAACTTGGCAGATATTCCATACAGTGTGGTGTCTTTTGAAAAAAAAGACTTCAAAATTGGTGGACCAGTTTCAAAACACTGCCAGAAATTAAAAACTACAATACCAAATGATGATAAATTGAAATCTCTAATAGAAGATAGTTTAAAATTACGATTGAATCAAGTAAATTTGTCTGCTGAGGTAAATTCACAGAGAGAATTGTTAAAAACTACAATTTTTGATTTTAGTTTTCTTAATGAATGTAAAGAATAG
Protein
MNDIVFVGDWIKDRVLGSGSFGTVVLWKHKKTEEKLAIKTCKWGDELTDKHRDRWTKEVEMLQSCRNPNIVGSKPLPSEFIKGLERGNPSGLPIMCMEYCSGGDLRQQLNKPESCSGLREIQVRQILSDIRNAMKFLHQKKITHRDLKPENIVMNIVENTEQNNISPQTEPRVIYKLIDLGYAKEIDFNSVCASFVGTLQYLAPELLYSKTYSNSVDFWSFGLLAFEIICGKRPFLPFMSPAQWMPHVEKKTHENICVYETFHGDIEYSNNIFPENHMSAPFKVLMEDWLRIALEWDPKLRGRDSHTRVRFNIPTEQREVSSNIIIYDLLENILSRKIIKIFSVPTLSRLAYQINDTTMVSDLKKWIGQDTSMDIADLILICPNTFTELGEMDIVSNYWREHCNTMVYLFNKTYILQKNIEPEVPKSVQRCLEHPKALYNHKNSQTLYKNSLYFVKNQSEIYESLINGLLVRGESLKNEGKQLLLKHNAVDKSISNLLAKEEIMKRLIETGKEQVNKLRDSGKGINMLGGFNKTFQDTDEHFLKISKLQQAWNQLSVRLTSAKRRCNEIVSDDIKNFVAKYNYQAVYANAFKIYIMARKSEIIGKEKHCDDIMKICYNCLKLRSKILLEIHHQPFVVKLMDLGTEFSKISDIVTKAAENTEKLTKDLSDYIDDLNNCIWSTINLAVSDGDNLADIPYSVVSFEKKDFKIGGPVSKHCQKLKTTIPNDDKLKSLIEDSLKLRLNQVNLSAEVNSQRELLKTTIFDFSFLNECKE
Summary
Uniprot
H9JFU2
A0A0L7L685
A0A2A4J527
A0A2H1W407
A0A0N0PCH1
A0A194PCC0
+ More
A0A067QNY8 A0A1B6IRA4 A0A2J7QMP3 A0A1L8DKU4 A0A1L8DKR3 A0A2P1AAT5 A0A0N7ZBM3 U5ERB4 A0A1S4F432 A0A1B0DJL5 A0A182GYE3 G5CJV9 A0A023EW54 A0A336LR16 Q17G02 A0A182SDM2 A0A182F1M6 A0A1B6DI45 A0A2M4A6A9 W5JHY4 A0A2M4A6F0 A0A182JYG1 A0A182Q3C8 A0A1Q3FFN9 A0A1Q3FFT1 A0A2M4A6D5 B0X7N9 A0A2M4A6D7 A0A182NF39 A0A2M4BE83 A0A2M4BE46 A0A182Y9R6 A0A182PFR0 A0A182MF39 A0A084VMW1 A0A1Y1M0J1 A0A1Y1LUR1 A0A182R6J6 A0A0D3QBW2 A0A0D3QCL5 A0A182WH61 A0A0D3QC04 A0A0D3QBW0 A0A0D3QBW4 A0A0D3QCF8 A0A0D3QCM4 Q5TNB7 A0A182WYL3 A0A0D3QC16 A0A0D3QC21 A0A182I557 A0A0D3QBV9 A0A0D3QCF4 A0A182UXR1 A0A182UF63 A0A1S4H136 A0A0D3QBV6 A0A0D3QBV5 A0A0D3QBU8 A0A0D3QCM0 A0A0D3QBV2 A0A182KUF9 A0A0D3QC12 A0A0D3QCG6 A0A0D3QCL1 A0A0D3QCG2 A0A0D3QC09 A0A182G3B4 A0A023EWK1 A0A210Q9S1
A0A067QNY8 A0A1B6IRA4 A0A2J7QMP3 A0A1L8DKU4 A0A1L8DKR3 A0A2P1AAT5 A0A0N7ZBM3 U5ERB4 A0A1S4F432 A0A1B0DJL5 A0A182GYE3 G5CJV9 A0A023EW54 A0A336LR16 Q17G02 A0A182SDM2 A0A182F1M6 A0A1B6DI45 A0A2M4A6A9 W5JHY4 A0A2M4A6F0 A0A182JYG1 A0A182Q3C8 A0A1Q3FFN9 A0A1Q3FFT1 A0A2M4A6D5 B0X7N9 A0A2M4A6D7 A0A182NF39 A0A2M4BE83 A0A2M4BE46 A0A182Y9R6 A0A182PFR0 A0A182MF39 A0A084VMW1 A0A1Y1M0J1 A0A1Y1LUR1 A0A182R6J6 A0A0D3QBW2 A0A0D3QCL5 A0A182WH61 A0A0D3QC04 A0A0D3QBW0 A0A0D3QBW4 A0A0D3QCF8 A0A0D3QCM4 Q5TNB7 A0A182WYL3 A0A0D3QC16 A0A0D3QC21 A0A182I557 A0A0D3QBV9 A0A0D3QCF4 A0A182UXR1 A0A182UF63 A0A1S4H136 A0A0D3QBV6 A0A0D3QBV5 A0A0D3QBU8 A0A0D3QCM0 A0A0D3QBV2 A0A182KUF9 A0A0D3QC12 A0A0D3QCG6 A0A0D3QCL1 A0A0D3QCG2 A0A0D3QC09 A0A182G3B4 A0A023EWK1 A0A210Q9S1
Pubmed
EMBL
BABH01001603
JTDY01002746
KOB70799.1
NWSH01003334
PCG66530.1
ODYU01006164
+ More
SOQ47778.1 KQ460556 KPJ14004.1 KQ459606 KPI90926.1 KK853183 KDR10135.1 GECU01018252 JAS89454.1 NEVH01013204 PNF29865.1 GFDF01007120 JAV06964.1 GFDF01007119 JAV06965.1 MF374338 AVI10168.1 GDRN01081265 JAI62048.1 GANO01002876 JAB56995.1 AJVK01065486 JXUM01097358 JXUM01097359 KQ564337 KXJ72408.1 JN180642 AEK86518.1 GAPW01000247 JAC13351.1 UFQT01000118 SSX20400.1 CH477267 EAT45468.1 GEDC01011942 JAS25356.1 GGFK01002970 MBW36291.1 ADMH02001355 ETN62893.1 GGFK01002981 MBW36302.1 AXCN02000829 GFDL01008684 JAV26361.1 GFDL01008641 JAV26404.1 GGFK01003046 MBW36367.1 DS232456 EDS42051.1 GGFK01002961 MBW36282.1 GGFJ01002186 MBW51327.1 GGFJ01002185 MBW51326.1 AXCM01014086 ATLV01014631 KE524975 KFB39305.1 GEZM01046260 JAV77376.1 GEZM01046266 JAV77372.1 KP274254 AJC98338.1 KP274250 AJC98334.1 KP274243 AJC98327.1 KP274262 AJC98346.1 KP274259 AJC98343.1 KP274251 AJC98335.1 KP274260 AJC98344.1 AAAB01008984 EAL39059.3 KP274258 AJC98342.1 KP274263 AJC98347.1 APCN01003784 KP274249 AJC98333.1 KP274246 AJC98330.1 KP274257 AJC98341.1 KP274244 AJC98328.1 KP274247 AJC98331.1 KP274255 AJC98339.1 KP274252 AJC98336.1 KP274253 AJC98337.1 KP274261 AJC98345.1 KP274245 AJC98329.1 KP274256 AJC98340.1 KP274248 AJC98332.1 JXUM01141228 KQ569208 KXJ68723.1 GBBI01005007 JAC13705.1 NEDP02004497 OWF45490.1
SOQ47778.1 KQ460556 KPJ14004.1 KQ459606 KPI90926.1 KK853183 KDR10135.1 GECU01018252 JAS89454.1 NEVH01013204 PNF29865.1 GFDF01007120 JAV06964.1 GFDF01007119 JAV06965.1 MF374338 AVI10168.1 GDRN01081265 JAI62048.1 GANO01002876 JAB56995.1 AJVK01065486 JXUM01097358 JXUM01097359 KQ564337 KXJ72408.1 JN180642 AEK86518.1 GAPW01000247 JAC13351.1 UFQT01000118 SSX20400.1 CH477267 EAT45468.1 GEDC01011942 JAS25356.1 GGFK01002970 MBW36291.1 ADMH02001355 ETN62893.1 GGFK01002981 MBW36302.1 AXCN02000829 GFDL01008684 JAV26361.1 GFDL01008641 JAV26404.1 GGFK01003046 MBW36367.1 DS232456 EDS42051.1 GGFK01002961 MBW36282.1 GGFJ01002186 MBW51327.1 GGFJ01002185 MBW51326.1 AXCM01014086 ATLV01014631 KE524975 KFB39305.1 GEZM01046260 JAV77376.1 GEZM01046266 JAV77372.1 KP274254 AJC98338.1 KP274250 AJC98334.1 KP274243 AJC98327.1 KP274262 AJC98346.1 KP274259 AJC98343.1 KP274251 AJC98335.1 KP274260 AJC98344.1 AAAB01008984 EAL39059.3 KP274258 AJC98342.1 KP274263 AJC98347.1 APCN01003784 KP274249 AJC98333.1 KP274246 AJC98330.1 KP274257 AJC98341.1 KP274244 AJC98328.1 KP274247 AJC98331.1 KP274255 AJC98339.1 KP274252 AJC98336.1 KP274253 AJC98337.1 KP274261 AJC98345.1 KP274245 AJC98329.1 KP274256 AJC98340.1 KP274248 AJC98332.1 JXUM01141228 KQ569208 KXJ68723.1 GBBI01005007 JAC13705.1 NEDP02004497 OWF45490.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000027135
+ More
UP000235965 UP000092462 UP000069940 UP000249989 UP000008820 UP000075901 UP000069272 UP000000673 UP000075881 UP000075886 UP000002320 UP000075884 UP000076408 UP000075885 UP000075883 UP000030765 UP000075900 UP000075920 UP000007062 UP000076407 UP000075840 UP000075903 UP000075902 UP000075882 UP000242188
UP000235965 UP000092462 UP000069940 UP000249989 UP000008820 UP000075901 UP000069272 UP000000673 UP000075881 UP000075886 UP000002320 UP000075884 UP000076408 UP000075885 UP000075883 UP000030765 UP000075900 UP000075920 UP000007062 UP000076407 UP000075840 UP000075903 UP000075902 UP000075882 UP000242188
PRIDE
Interpro
ProteinModelPortal
H9JFU2
A0A0L7L685
A0A2A4J527
A0A2H1W407
A0A0N0PCH1
A0A194PCC0
+ More
A0A067QNY8 A0A1B6IRA4 A0A2J7QMP3 A0A1L8DKU4 A0A1L8DKR3 A0A2P1AAT5 A0A0N7ZBM3 U5ERB4 A0A1S4F432 A0A1B0DJL5 A0A182GYE3 G5CJV9 A0A023EW54 A0A336LR16 Q17G02 A0A182SDM2 A0A182F1M6 A0A1B6DI45 A0A2M4A6A9 W5JHY4 A0A2M4A6F0 A0A182JYG1 A0A182Q3C8 A0A1Q3FFN9 A0A1Q3FFT1 A0A2M4A6D5 B0X7N9 A0A2M4A6D7 A0A182NF39 A0A2M4BE83 A0A2M4BE46 A0A182Y9R6 A0A182PFR0 A0A182MF39 A0A084VMW1 A0A1Y1M0J1 A0A1Y1LUR1 A0A182R6J6 A0A0D3QBW2 A0A0D3QCL5 A0A182WH61 A0A0D3QC04 A0A0D3QBW0 A0A0D3QBW4 A0A0D3QCF8 A0A0D3QCM4 Q5TNB7 A0A182WYL3 A0A0D3QC16 A0A0D3QC21 A0A182I557 A0A0D3QBV9 A0A0D3QCF4 A0A182UXR1 A0A182UF63 A0A1S4H136 A0A0D3QBV6 A0A0D3QBV5 A0A0D3QBU8 A0A0D3QCM0 A0A0D3QBV2 A0A182KUF9 A0A0D3QC12 A0A0D3QCG6 A0A0D3QCL1 A0A0D3QCG2 A0A0D3QC09 A0A182G3B4 A0A023EWK1 A0A210Q9S1
A0A067QNY8 A0A1B6IRA4 A0A2J7QMP3 A0A1L8DKU4 A0A1L8DKR3 A0A2P1AAT5 A0A0N7ZBM3 U5ERB4 A0A1S4F432 A0A1B0DJL5 A0A182GYE3 G5CJV9 A0A023EW54 A0A336LR16 Q17G02 A0A182SDM2 A0A182F1M6 A0A1B6DI45 A0A2M4A6A9 W5JHY4 A0A2M4A6F0 A0A182JYG1 A0A182Q3C8 A0A1Q3FFN9 A0A1Q3FFT1 A0A2M4A6D5 B0X7N9 A0A2M4A6D7 A0A182NF39 A0A2M4BE83 A0A2M4BE46 A0A182Y9R6 A0A182PFR0 A0A182MF39 A0A084VMW1 A0A1Y1M0J1 A0A1Y1LUR1 A0A182R6J6 A0A0D3QBW2 A0A0D3QCL5 A0A182WH61 A0A0D3QC04 A0A0D3QBW0 A0A0D3QBW4 A0A0D3QCF8 A0A0D3QCM4 Q5TNB7 A0A182WYL3 A0A0D3QC16 A0A0D3QC21 A0A182I557 A0A0D3QBV9 A0A0D3QCF4 A0A182UXR1 A0A182UF63 A0A1S4H136 A0A0D3QBV6 A0A0D3QBV5 A0A0D3QBU8 A0A0D3QCM0 A0A0D3QBV2 A0A182KUF9 A0A0D3QC12 A0A0D3QCG6 A0A0D3QCL1 A0A0D3QCG2 A0A0D3QC09 A0A182G3B4 A0A023EWK1 A0A210Q9S1
PDB
3QA8
E-value=1.04546e-80,
Score=767
Ontologies
PATHWAY
GO
Topology
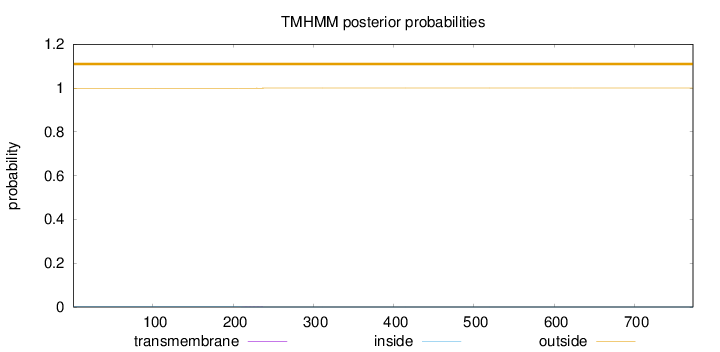
Length:
773
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01647
Exp number, first 60 AAs:
0.00221
Total prob of N-in:
0.00075
outside
1 - 773
Population Genetic Test Statistics
Pi
275.707601
Theta
237.513578
Tajima's D
0.528743
CLR
0.005114
CSRT
0.520173991300435
Interpretation
Uncertain
