Gene
KWMTBOMO10784
Pre Gene Modal
BGIBMGA008428
Annotation
Uncharacterized_protein_OBRU01_14771_[Operophtera_brumata]
Full name
Minichromosome maintenance domain-containing protein 2
Location in the cell
PlasmaMembrane Reliability : 2.779
Sequence
CDS
ATGGAGCTGCATTGTGAATTATTGCTATATTTAGATAAAAGGCGCTATTTTGCTAAAATGAAGGATGATTGTGAAAATTTCTTAGAATCATTAACAGAAAAAACAGTGAGTAAGTTTCCTTCACTGCGATGTTTCCTAGAAATTGACGTCATGGATGTAATCGACGTGTTGCCGAGTCTTGGAGAATTAATAATAAAAGAACCATTGAAATTGCAAAAGATGTGCAATGACGTATTATTTGCGTGTCTGCGATCTATTGACGTGGATAACTACAAGCTTGTTGAATATTCACAAGTTGTCGTTGTTTTAAGGTTAAAATGTGTTCCTCGAATTCTATGCCTACCCAACCCAAAACGGTATAAAGGCTTAACGTATTTTGAAGGGACATTATTAGCTAAATCAGAATCTGTAAGCTATGTATATCACACAGTCTGGTCATGTCCGGAAGATTGTGAAGACAGCATAAACATAATACATTATATACCAAAAACACCACCAAAATGTTGCATTTGTAGAAGTACAATGTACGAGAACAGCGGCCTCAGAAGATGCGGCGACCAAGTTACAGCAACTTTCAAATTGAAAGGAGACTATTTATTAAGAAAATTCTACATAGTCGATGATTTAATTCCCAAATTAAAAGTGGGTTTATATTACGCCTTGCATGCTATTGTAGGTAAATCCATTATAATATGGAGTTTGGAAAAGATTGAACAATGTAAAGCACCAATGACCCATCCCGTGCCCAGGGACTTGGATGTATTGTATAACGCATGCAATCGTTCCCCTTGGAAATTCATATATTGTCTCGCTTCCAGTCTAGCGATTAACGTGTGTCCTTTAAACTGCTTCATACAATTGAAGATAAATTTATTATTGAGTCTAGCAAGCATTAAAGCTAACGTACTGACTAATTCTAATATCCTACACGTACTAGCAACAGGATTTGATACTCGTTTTGTGGGAGAAATTATGAATGAAGCGGCTAAGCTTACGGATAAAAATATTTTAATAGGAACTTCTAATACTGATGTTGCCACTGCTTTGATCGCGGGTACTGGAGGAATAGCCGCATTTCCACTACCACTTCACGCTTACAATCAAAAACAAATATTTTCTGTTCTATCAGCCATTGAAAGTGGAGAAATAATGAATGAGACAAGTAAAACAAAACTGAAATGTGCTGTTTGGGCGCATGGAATGGATTTTAAAAAGATCGTAATGTTCAATGTTGCTAGTGTATTTGGCAATGTGTGTAGAGGTGACCACGGCGAATTCAATGATAATATTGTAGAGTATTTATTACAAGGATCCATAGAACCTTCCGGAATAACAGAAGATGAACTACGTGCACTGAAAGATATTAGCACATATTTAAGCATTGTGGCTGGCATAGAAGTTGAATTAGATAAATTACCAGAAACACTTCTGAGAAGCTATTTTCTTGCAGCTCGTAAGGAAAGACCTCGTGCAGTTGCGATTAGCAGTATGAACTCGCTAATCACTGTTAGCCTTACATCGGCGAGGCTTTGCAGGCGCTCAGTTGTTACTATAGAAGACGCTATATTTGCTATATGGCTGCATGTATGTGGATTTCCAGAGCCTAGATTCGCCCCGGAGGAATATCTTGAAACTCCAATGAATATCCGAAAGTTGAACAAGATCATGGACGGCTTCAAACATTGGCTCGAACAGTTTACTGGGATGAGTAATTTGTAA
Protein
MELHCELLLYLDKRRYFAKMKDDCENFLESLTEKTVSKFPSLRCFLEIDVMDVIDVLPSLGELIIKEPLKLQKMCNDVLFACLRSIDVDNYKLVEYSQVVVVLRLKCVPRILCLPNPKRYKGLTYFEGTLLAKSESVSYVYHTVWSCPEDCEDSINIIHYIPKTPPKCCICRSTMYENSGLRRCGDQVTATFKLKGDYLLRKFYIVDDLIPKLKVGLYYALHAIVGKSIIIWSLEKIEQCKAPMTHPVPRDLDVLYNACNRSPWKFIYCLASSLAINVCPLNCFIQLKINLLLSLASIKANVLTNSNILHVLATGFDTRFVGEIMNEAAKLTDKNILIGTSNTDVATALIAGTGGIAAFPLPLHAYNQKQIFSVLSAIESGEIMNETSKTKLKCAVWAHGMDFKKIVMFNVASVFGNVCRGDHGEFNDNIVEYLLQGSIEPSGITEDELRALKDISTYLSIVAGIEVELDKLPETLLRSYFLAARKERPRAVAISSMNSLITVSLTSARLCRRSVVTIEDAIFAIWLHVCGFPEPRFAPEEYLETPMNIRKLNKIMDGFKHWLEQFTGMSNL
Summary
Description
Plays an important role in meiotic recombination and associated DNA double-strand break repair.
Keywords
Alternative splicing
Complete proteome
DNA damage
DNA repair
Meiosis
Phosphoprotein
Reference proteome
Feature
chain Minichromosome maintenance domain-containing protein 2
splice variant In isoform 2.
splice variant In isoform 2.
Uniprot
H9JFY1
A0A2A4J3G7
A0A2H1W3V3
A0A0L7L655
A0A194PCK2
A0A212FC54
+ More
A0A1B6G867 A0A1B6EYN5 H2TFH4 A0A3P9JGM3 A0A224XJV2 A0A3P9L0B8 A0A3S2NX71 F6RB54 H3CXL9 A0A3P9JFV2 A0A3Q0SVZ2 A0A3B4UL46 W5L2N5 A0A1B6CLK7 A0A3Q1JQB0 A0A1L8FZ43 A0A3Q1D083 A0A3Q3IK26 A0A3B4UM48 H2LWS7 A0A3P8W8G8 I3K9Q5 F6ZS93 A0A3B3YMC3 E6ZIQ0 A0A3P8RA62 A0A091NPJ6 A0A1V4KGS1 A0A151MNB9 A0A3P9D3M3 A0A091S883 A0A091QA66 A0A093H5Z8 A0A3B3D9B3 A0A1U7S440 A0A3Q3C998 A0A3B4F1C3 A0A3B3VCA6 A0A091VP81 A0A093HFY0 G1Q7N4 E9Q956 R9PXX4 Q5XI14 H7BXC1 A0A2J8UR66 A0A3Q3FLH5 A0A091IXU6 A0A093QNS6 A0A091E779 A0A094KWA4 A0A2J8UR73 A0A3P9Q703 A0A087QI41 A0A091L686 A0A0L8GTL8 A0A093J1K8 A0A1V4KGG9 G1NFI2 A0A3Q3W6V7 A0A093NDS9 A0A0Q3P6D8 A0A3P8TSS3 A0A093F6U8 A0A087XEZ8 K7G5W8 A0A3Q4I3A4 A0A151MN93 A0A1S2ZZS3 G1TUL7 A0A1W4Z7C6 A0A3Q3IAU5 A0A0P7VHR3 A0A151MN95 U3JSA4 A0A3B4BI61 A0A1S3QA92 A0A3Q3T1D2 A0A3Q2D8Q8 A0A091WIJ3 A0A099ZLF6 A0A2K5C5B1 A0A2Y9GY47 A0A091TSX7 H0ZM52 A0A2U3XSR4 A0A060XJI0 A0A3Q7NZ04 A0A2K6T5Q7 M3ZH92 W5Q932 A0A2D0PSA4
A0A1B6G867 A0A1B6EYN5 H2TFH4 A0A3P9JGM3 A0A224XJV2 A0A3P9L0B8 A0A3S2NX71 F6RB54 H3CXL9 A0A3P9JFV2 A0A3Q0SVZ2 A0A3B4UL46 W5L2N5 A0A1B6CLK7 A0A3Q1JQB0 A0A1L8FZ43 A0A3Q1D083 A0A3Q3IK26 A0A3B4UM48 H2LWS7 A0A3P8W8G8 I3K9Q5 F6ZS93 A0A3B3YMC3 E6ZIQ0 A0A3P8RA62 A0A091NPJ6 A0A1V4KGS1 A0A151MNB9 A0A3P9D3M3 A0A091S883 A0A091QA66 A0A093H5Z8 A0A3B3D9B3 A0A1U7S440 A0A3Q3C998 A0A3B4F1C3 A0A3B3VCA6 A0A091VP81 A0A093HFY0 G1Q7N4 E9Q956 R9PXX4 Q5XI14 H7BXC1 A0A2J8UR66 A0A3Q3FLH5 A0A091IXU6 A0A093QNS6 A0A091E779 A0A094KWA4 A0A2J8UR73 A0A3P9Q703 A0A087QI41 A0A091L686 A0A0L8GTL8 A0A093J1K8 A0A1V4KGG9 G1NFI2 A0A3Q3W6V7 A0A093NDS9 A0A0Q3P6D8 A0A3P8TSS3 A0A093F6U8 A0A087XEZ8 K7G5W8 A0A3Q4I3A4 A0A151MN93 A0A1S2ZZS3 G1TUL7 A0A1W4Z7C6 A0A3Q3IAU5 A0A0P7VHR3 A0A151MN95 U3JSA4 A0A3B4BI61 A0A1S3QA92 A0A3Q3T1D2 A0A3Q2D8Q8 A0A091WIJ3 A0A099ZLF6 A0A2K5C5B1 A0A2Y9GY47 A0A091TSX7 H0ZM52 A0A2U3XSR4 A0A060XJI0 A0A3Q7NZ04 A0A2K6T5Q7 M3ZH92 W5Q932 A0A2D0PSA4
Pubmed
EMBL
BABH01001602
NWSH01003334
PCG66531.1
ODYU01006164
SOQ47779.1
JTDY01002746
+ More
KOB70796.1 KQ459606 KPI90927.1 AGBW02009236 OWR51297.1 GECZ01011188 JAS58581.1 GECZ01026818 JAS42951.1 GFTR01007544 JAW08882.1 CM012456 RVE58752.1 AAMC01053906 GEDC01023030 JAS14268.1 CM004476 OCT76858.1 AERX01010045 AAPN01326410 AAPN01326411 FQ310508 CBN81934.1 KL389212 KFP91027.1 LSYS01003169 OPJ83533.1 AKHW03005657 KYO26006.1 KK943473 KFQ52508.1 KK672866 KFQ06479.1 KL206020 KFV78013.1 KL411206 KFR04450.1 KK389476 KFV53548.1 AAPE02021717 AK031395 AC102570 AABR07046831 AABR07046832 AABR07046833 AABR07046834 BC083882 NDHI03003448 PNJ47764.1 KK501045 KFP12393.1 KL725410 KFW88040.1 KK717901 KFO53026.1 KL346217 KFZ55975.1 PNJ47767.1 KL225613 KFM00895.1 KL305238 KFP51994.1 KQ420424 KOF80323.1 KK571667 KFW07798.1 OPJ83532.1 KL224729 KFW63063.1 LMAW01002862 KQK76331.1 KK626069 KFV53382.1 AYCK01005747 AGCU01142028 KYO26007.1 AAGW02046995 JARO02002301 KPP73026.1 KYO26008.1 AGTO01001016 KK735701 KFR15384.1 KL895340 KGL82577.1 KK463329 KFQ80453.1 ABQF01005320 FR905481 CDQ79626.1 AMGL01111303 AMGL01111304 AMGL01111305 AMGL01111306
KOB70796.1 KQ459606 KPI90927.1 AGBW02009236 OWR51297.1 GECZ01011188 JAS58581.1 GECZ01026818 JAS42951.1 GFTR01007544 JAW08882.1 CM012456 RVE58752.1 AAMC01053906 GEDC01023030 JAS14268.1 CM004476 OCT76858.1 AERX01010045 AAPN01326410 AAPN01326411 FQ310508 CBN81934.1 KL389212 KFP91027.1 LSYS01003169 OPJ83533.1 AKHW03005657 KYO26006.1 KK943473 KFQ52508.1 KK672866 KFQ06479.1 KL206020 KFV78013.1 KL411206 KFR04450.1 KK389476 KFV53548.1 AAPE02021717 AK031395 AC102570 AABR07046831 AABR07046832 AABR07046833 AABR07046834 BC083882 NDHI03003448 PNJ47764.1 KK501045 KFP12393.1 KL725410 KFW88040.1 KK717901 KFO53026.1 KL346217 KFZ55975.1 PNJ47767.1 KL225613 KFM00895.1 KL305238 KFP51994.1 KQ420424 KOF80323.1 KK571667 KFW07798.1 OPJ83532.1 KL224729 KFW63063.1 LMAW01002862 KQK76331.1 KK626069 KFV53382.1 AYCK01005747 AGCU01142028 KYO26007.1 AAGW02046995 JARO02002301 KPP73026.1 KYO26008.1 AGTO01001016 KK735701 KFR15384.1 KL895340 KGL82577.1 KK463329 KFQ80453.1 ABQF01005320 FR905481 CDQ79626.1 AMGL01111303 AMGL01111304 AMGL01111305 AMGL01111306
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000007151
UP000005226
+ More
UP000265200 UP000265180 UP000008143 UP000007303 UP000261340 UP000261420 UP000018467 UP000265040 UP000186698 UP000257160 UP000261600 UP000001038 UP000265120 UP000005207 UP000002279 UP000261480 UP000265100 UP000190648 UP000050525 UP000265160 UP000053584 UP000261560 UP000189705 UP000264840 UP000261460 UP000261500 UP000053283 UP000001074 UP000000589 UP000002494 UP000261660 UP000053119 UP000053258 UP000052976 UP000242638 UP000053286 UP000053454 UP000001645 UP000261620 UP000054081 UP000051836 UP000265080 UP000028760 UP000007267 UP000261580 UP000079721 UP000001811 UP000192224 UP000034805 UP000016665 UP000261520 UP000087266 UP000261640 UP000265020 UP000053605 UP000053641 UP000233020 UP000248481 UP000007754 UP000245341 UP000193380 UP000286641 UP000233220 UP000002852 UP000002356 UP000221080
UP000265200 UP000265180 UP000008143 UP000007303 UP000261340 UP000261420 UP000018467 UP000265040 UP000186698 UP000257160 UP000261600 UP000001038 UP000265120 UP000005207 UP000002279 UP000261480 UP000265100 UP000190648 UP000050525 UP000265160 UP000053584 UP000261560 UP000189705 UP000264840 UP000261460 UP000261500 UP000053283 UP000001074 UP000000589 UP000002494 UP000261660 UP000053119 UP000053258 UP000052976 UP000242638 UP000053286 UP000053454 UP000001645 UP000261620 UP000054081 UP000051836 UP000265080 UP000028760 UP000007267 UP000261580 UP000079721 UP000001811 UP000192224 UP000034805 UP000016665 UP000261520 UP000087266 UP000261640 UP000265020 UP000053605 UP000053641 UP000233020 UP000248481 UP000007754 UP000245341 UP000193380 UP000286641 UP000233220 UP000002852 UP000002356 UP000221080
Interpro
ProteinModelPortal
H9JFY1
A0A2A4J3G7
A0A2H1W3V3
A0A0L7L655
A0A194PCK2
A0A212FC54
+ More
A0A1B6G867 A0A1B6EYN5 H2TFH4 A0A3P9JGM3 A0A224XJV2 A0A3P9L0B8 A0A3S2NX71 F6RB54 H3CXL9 A0A3P9JFV2 A0A3Q0SVZ2 A0A3B4UL46 W5L2N5 A0A1B6CLK7 A0A3Q1JQB0 A0A1L8FZ43 A0A3Q1D083 A0A3Q3IK26 A0A3B4UM48 H2LWS7 A0A3P8W8G8 I3K9Q5 F6ZS93 A0A3B3YMC3 E6ZIQ0 A0A3P8RA62 A0A091NPJ6 A0A1V4KGS1 A0A151MNB9 A0A3P9D3M3 A0A091S883 A0A091QA66 A0A093H5Z8 A0A3B3D9B3 A0A1U7S440 A0A3Q3C998 A0A3B4F1C3 A0A3B3VCA6 A0A091VP81 A0A093HFY0 G1Q7N4 E9Q956 R9PXX4 Q5XI14 H7BXC1 A0A2J8UR66 A0A3Q3FLH5 A0A091IXU6 A0A093QNS6 A0A091E779 A0A094KWA4 A0A2J8UR73 A0A3P9Q703 A0A087QI41 A0A091L686 A0A0L8GTL8 A0A093J1K8 A0A1V4KGG9 G1NFI2 A0A3Q3W6V7 A0A093NDS9 A0A0Q3P6D8 A0A3P8TSS3 A0A093F6U8 A0A087XEZ8 K7G5W8 A0A3Q4I3A4 A0A151MN93 A0A1S2ZZS3 G1TUL7 A0A1W4Z7C6 A0A3Q3IAU5 A0A0P7VHR3 A0A151MN95 U3JSA4 A0A3B4BI61 A0A1S3QA92 A0A3Q3T1D2 A0A3Q2D8Q8 A0A091WIJ3 A0A099ZLF6 A0A2K5C5B1 A0A2Y9GY47 A0A091TSX7 H0ZM52 A0A2U3XSR4 A0A060XJI0 A0A3Q7NZ04 A0A2K6T5Q7 M3ZH92 W5Q932 A0A2D0PSA4
A0A1B6G867 A0A1B6EYN5 H2TFH4 A0A3P9JGM3 A0A224XJV2 A0A3P9L0B8 A0A3S2NX71 F6RB54 H3CXL9 A0A3P9JFV2 A0A3Q0SVZ2 A0A3B4UL46 W5L2N5 A0A1B6CLK7 A0A3Q1JQB0 A0A1L8FZ43 A0A3Q1D083 A0A3Q3IK26 A0A3B4UM48 H2LWS7 A0A3P8W8G8 I3K9Q5 F6ZS93 A0A3B3YMC3 E6ZIQ0 A0A3P8RA62 A0A091NPJ6 A0A1V4KGS1 A0A151MNB9 A0A3P9D3M3 A0A091S883 A0A091QA66 A0A093H5Z8 A0A3B3D9B3 A0A1U7S440 A0A3Q3C998 A0A3B4F1C3 A0A3B3VCA6 A0A091VP81 A0A093HFY0 G1Q7N4 E9Q956 R9PXX4 Q5XI14 H7BXC1 A0A2J8UR66 A0A3Q3FLH5 A0A091IXU6 A0A093QNS6 A0A091E779 A0A094KWA4 A0A2J8UR73 A0A3P9Q703 A0A087QI41 A0A091L686 A0A0L8GTL8 A0A093J1K8 A0A1V4KGG9 G1NFI2 A0A3Q3W6V7 A0A093NDS9 A0A0Q3P6D8 A0A3P8TSS3 A0A093F6U8 A0A087XEZ8 K7G5W8 A0A3Q4I3A4 A0A151MN93 A0A1S2ZZS3 G1TUL7 A0A1W4Z7C6 A0A3Q3IAU5 A0A0P7VHR3 A0A151MN95 U3JSA4 A0A3B4BI61 A0A1S3QA92 A0A3Q3T1D2 A0A3Q2D8Q8 A0A091WIJ3 A0A099ZLF6 A0A2K5C5B1 A0A2Y9GY47 A0A091TSX7 H0ZM52 A0A2U3XSR4 A0A060XJI0 A0A3Q7NZ04 A0A2K6T5Q7 M3ZH92 W5Q932 A0A2D0PSA4
Ontologies
GO
PANTHER
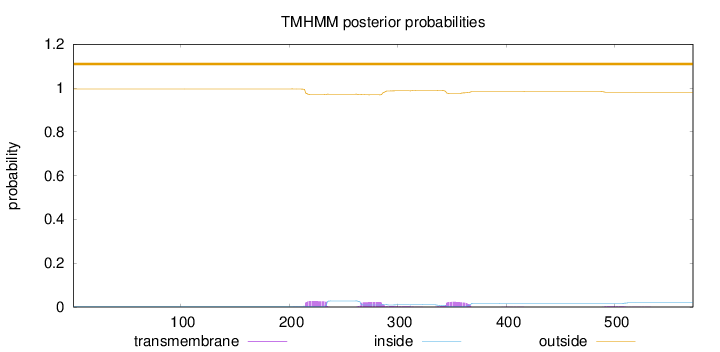
Topology
Length:
572
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.66262
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00396
outside
1 - 572
Population Genetic Test Statistics
Pi
207.398785
Theta
189.197905
Tajima's D
0.163855
CLR
0.010254
CSRT
0.414979251037448
Interpretation
Uncertain
