Gene
KWMTBOMO10781
Pre Gene Modal
BGIBMGA008427
Annotation
PREDICTED:_nuclear_nucleic_acid-binding_protein_C1D-like_[Amyelois_transitella]
Full name
Nuclear nucleic acid-binding protein C1D
Location in the cell
Mitochondrial Reliability : 1.512 Nuclear Reliability : 2.12
Sequence
CDS
ATGTGCCAAGAACTTAATTGGGACTTTAAATACGGCGAACTCGCCAAAGACAAAGATTTCGTGAACAATGTTGAAAATTTGAAAGAGAATTTAATAGAAGTACAACAAGTACTCGATAAACTCCTTCCGTTGAAGAAAAATTACGACAAAATGTCACTACCAGCTCAGATTGAACTAGATTTGTTTTTTGTATACACTTTAAATTCACTACATTGGATACATTTACGAACAAAAGGCATCGATCCCACAAAACATCCCATCAAAGATGAACTGCTCAGAATAAAAGCTACAATGTTAAAGTGGCAGGAAGTGAAGGATCGCCAGAAAAGACCCACCGTTAACGTTGAAGTGGCGAAACGTTTAGTACGGAATGGTCTGTACGACCATCAGAGGGCTCCCGTGAAACAACTAAACAAACGAATAAAATTTTCAGATAACGAAGAGTAA
Protein
MCQELNWDFKYGELAKDKDFVNNVENLKENLIEVQQVLDKLLPLKKNYDKMSLPAQIELDLFFVYTLNSLHWIHLRTKGIDPTKHPIKDELLRIKATMLKWQEVKDRQKRPTVNVEVAKRLVRNGLYDHQRAPVKQLNKRIKFSDNEE
Summary
Description
Plays a role in the recruitment of the exosome to pre-rRNA to mediate the 3'-5' end processing of the 5.8S rRNA.
Subunit
Monomer and homodimer.
Similarity
Belongs to the C1D family.
Keywords
Coiled coil
Cytoplasm
DNA-binding
Exosome
Nucleus
RNA-binding
rRNA processing
Complete proteome
Reference proteome
Feature
chain Nuclear nucleic acid-binding protein C1D
Uniprot
H9JFY0
A0A2A4J4F8
A0A3S2LRD3
A0A1E1W4P0
A0A2H1VZ27
A0A0L7LJ51
+ More
A0A212ETP3 S4P8H6 A0A0N1IG38 A0A194PIL3 A0A023EH30 Q17GP9 A0A1Q3FCB8 B0XJI0 A0A0A9WDV8 A0A1L8DCD1 U5EN26 T1GS34 A0A1J1IDS1 D6WVJ5 A0A023EZ69 A0A161N176 A0A2J7QAZ1 T1I8V0 A0A2P8Y2H6 E0VG57 J3JV10 A0A1I8PLW8 A0A0P4VRG2 A0A224XLE5 A0A1B0D5Z2 K7J7U0 A0A1L8EC75 W8BR41 A0A1I8MEV9 A0A2R7VWM3 A0A067QSL4 A0A069DPF0 A0A067QT38 A0A1W4X785 C4WVB2 A0A154PCB6 A0A0A1XME6 A0A2S2NL90 A0A0K8V1N3 G3PE26 A0A026W7E7 A0A2A3E8F0 A0A1B6EKY4 A0A034WXW0 A0A0C9QEM9 A0A3P8UC68 A0A336LJI0 A0A3Q3B7H8 A0A088ABI5 A0A0L0CGD4 A0A0N0U307 A0A1B6LSD0 A0A3Q2FHJ9 C1C2U7 A0A1B0A3F1 A0A3Q2GBJ3 A0A0K8TMF7 A0A091RSK6 A0A2I0M982 A0A1A9YDF9 A0A1B0B009 C1C0D4 A0A2I0M985 A0A3B1K3D5 A0A2H8TU46 A0A1A8ECM6 A0A1L8G708 Q3KPR1 A0A1V4IG26 A0A3B4A2J3 A0A3P8UEL5 A0A1A8K4X6 A0A3B3IN79 A0A091M8P8 A0A091HF74 A0A3B5M4G4 A0A1A8QL70 A0A1A8HRK2 A0A093GLT7 A0A2S2R679 F1QNQ8 A0A1A8VDV5 A0A3P9IWE8 Q5XJ97
A0A212ETP3 S4P8H6 A0A0N1IG38 A0A194PIL3 A0A023EH30 Q17GP9 A0A1Q3FCB8 B0XJI0 A0A0A9WDV8 A0A1L8DCD1 U5EN26 T1GS34 A0A1J1IDS1 D6WVJ5 A0A023EZ69 A0A161N176 A0A2J7QAZ1 T1I8V0 A0A2P8Y2H6 E0VG57 J3JV10 A0A1I8PLW8 A0A0P4VRG2 A0A224XLE5 A0A1B0D5Z2 K7J7U0 A0A1L8EC75 W8BR41 A0A1I8MEV9 A0A2R7VWM3 A0A067QSL4 A0A069DPF0 A0A067QT38 A0A1W4X785 C4WVB2 A0A154PCB6 A0A0A1XME6 A0A2S2NL90 A0A0K8V1N3 G3PE26 A0A026W7E7 A0A2A3E8F0 A0A1B6EKY4 A0A034WXW0 A0A0C9QEM9 A0A3P8UC68 A0A336LJI0 A0A3Q3B7H8 A0A088ABI5 A0A0L0CGD4 A0A0N0U307 A0A1B6LSD0 A0A3Q2FHJ9 C1C2U7 A0A1B0A3F1 A0A3Q2GBJ3 A0A0K8TMF7 A0A091RSK6 A0A2I0M982 A0A1A9YDF9 A0A1B0B009 C1C0D4 A0A2I0M985 A0A3B1K3D5 A0A2H8TU46 A0A1A8ECM6 A0A1L8G708 Q3KPR1 A0A1V4IG26 A0A3B4A2J3 A0A3P8UEL5 A0A1A8K4X6 A0A3B3IN79 A0A091M8P8 A0A091HF74 A0A3B5M4G4 A0A1A8QL70 A0A1A8HRK2 A0A093GLT7 A0A2S2R679 F1QNQ8 A0A1A8VDV5 A0A3P9IWE8 Q5XJ97
Pubmed
19121390
26227816
22118469
23622113
26354079
24945155
+ More
26483478 17510324 25401762 26823975 18362917 19820115 25474469 29403074 20566863 22516182 23537049 27129103 20075255 24495485 25315136 24845553 26334808 25830018 24508170 30249741 25348373 24487278 26108605 26369729 23371554 25329095 27762356 25463417 17554307 23594743
26483478 17510324 25401762 26823975 18362917 19820115 25474469 29403074 20566863 22516182 23537049 27129103 20075255 24495485 25315136 24845553 26334808 25830018 24508170 30249741 25348373 24487278 26108605 26369729 23371554 25329095 27762356 25463417 17554307 23594743
EMBL
BABH01001598
NWSH01003288
PCG66616.1
RSAL01000298
RVE42795.1
GDQN01009124
+ More
JAT81930.1 ODYU01005346 SOQ46095.1 JTDY01000907 KOB75477.1 AGBW02012556 OWR44837.1 GAIX01005926 JAA86634.1 KQ460556 KPJ14000.1 KQ459606 KPI90930.1 JXUM01057087 GAPW01005464 KQ561939 JAC08134.1 KXJ77104.1 CH477257 EAT45817.1 GFDL01009821 JAV25224.1 DS233521 EDS30383.1 GBHO01037623 GBHO01037620 GBHO01037618 GBHO01018799 GBHO01018798 GBRD01002861 GDHC01013226 GDHC01012111 JAG05981.1 JAG05984.1 JAG05986.1 JAG24805.1 JAG24806.1 JAG62960.1 JAQ05403.1 JAQ06518.1 GFDF01010057 JAV04027.1 GANO01000756 JAB59115.1 CAQQ02178077 CVRI01000044 CRK96593.1 KQ971357 EFA08579.1 GBBI01004200 JAC14512.1 GEMB01002371 JAS00812.1 NEVH01016304 PNF25768.1 ACPB03000011 PYGN01001014 PSN38468.1 DS235131 EEB12363.1 APGK01026888 BT127075 KB740648 KB630779 KB631357 AEE62037.1 ENN79721.1 ERL83865.1 ERL84254.1 GDKW01002967 JAI53628.1 GFTR01003139 JAW13287.1 AJVK01003541 AJVK01003542 GFDG01002449 JAV16350.1 GAMC01002790 JAC03766.1 KK854135 PTY11907.1 KK852984 KDR12859.1 GBGD01003392 JAC85497.1 KDR12860.1 ABLF02015467 ABLF02015468 AK341439 BAH71832.1 KQ434870 KZC09471.1 GBXI01002160 JAD12132.1 GGMR01005093 MBY17712.1 GDHF01025690 GDHF01022826 GDHF01019578 JAI26624.1 JAI29488.1 JAI32736.1 KK107356 QOIP01000010 EZA52027.1 RLU17432.1 KZ288347 PBC27552.1 GECZ01031208 JAS38561.1 GAKP01000329 JAC58623.1 GBYB01012958 JAG82725.1 UFQT01000027 SSX18162.1 JRES01000417 KNC31453.1 KQ435955 KOX68058.1 GEBQ01013375 JAT26602.1 BT081176 ACO15600.1 GDAI01002051 JAI15552.1 KK707790 KFQ31447.1 AKCR02000027 PKK26242.1 JXJN01006514 BT080313 ACO14737.1 PKK26240.1 PKK26241.1 GFXV01005922 MBW17727.1 HAEA01015198 SBQ43678.1 CM004474 OCT79678.1 BC106601 LSYS01009753 OPJ58810.1 HAEE01007246 SBR27266.1 KL321027 KFP54876.1 KL535088 KFO94119.1 HAEH01012150 SBR94063.1 HAED01000872 HAEE01010888 SBQ86717.1 KL216062 KFV67967.1 GGMS01015659 MBY84862.1 CABZ01017629 HAEJ01017589 SBS58046.1 BC083407
JAT81930.1 ODYU01005346 SOQ46095.1 JTDY01000907 KOB75477.1 AGBW02012556 OWR44837.1 GAIX01005926 JAA86634.1 KQ460556 KPJ14000.1 KQ459606 KPI90930.1 JXUM01057087 GAPW01005464 KQ561939 JAC08134.1 KXJ77104.1 CH477257 EAT45817.1 GFDL01009821 JAV25224.1 DS233521 EDS30383.1 GBHO01037623 GBHO01037620 GBHO01037618 GBHO01018799 GBHO01018798 GBRD01002861 GDHC01013226 GDHC01012111 JAG05981.1 JAG05984.1 JAG05986.1 JAG24805.1 JAG24806.1 JAG62960.1 JAQ05403.1 JAQ06518.1 GFDF01010057 JAV04027.1 GANO01000756 JAB59115.1 CAQQ02178077 CVRI01000044 CRK96593.1 KQ971357 EFA08579.1 GBBI01004200 JAC14512.1 GEMB01002371 JAS00812.1 NEVH01016304 PNF25768.1 ACPB03000011 PYGN01001014 PSN38468.1 DS235131 EEB12363.1 APGK01026888 BT127075 KB740648 KB630779 KB631357 AEE62037.1 ENN79721.1 ERL83865.1 ERL84254.1 GDKW01002967 JAI53628.1 GFTR01003139 JAW13287.1 AJVK01003541 AJVK01003542 GFDG01002449 JAV16350.1 GAMC01002790 JAC03766.1 KK854135 PTY11907.1 KK852984 KDR12859.1 GBGD01003392 JAC85497.1 KDR12860.1 ABLF02015467 ABLF02015468 AK341439 BAH71832.1 KQ434870 KZC09471.1 GBXI01002160 JAD12132.1 GGMR01005093 MBY17712.1 GDHF01025690 GDHF01022826 GDHF01019578 JAI26624.1 JAI29488.1 JAI32736.1 KK107356 QOIP01000010 EZA52027.1 RLU17432.1 KZ288347 PBC27552.1 GECZ01031208 JAS38561.1 GAKP01000329 JAC58623.1 GBYB01012958 JAG82725.1 UFQT01000027 SSX18162.1 JRES01000417 KNC31453.1 KQ435955 KOX68058.1 GEBQ01013375 JAT26602.1 BT081176 ACO15600.1 GDAI01002051 JAI15552.1 KK707790 KFQ31447.1 AKCR02000027 PKK26242.1 JXJN01006514 BT080313 ACO14737.1 PKK26240.1 PKK26241.1 GFXV01005922 MBW17727.1 HAEA01015198 SBQ43678.1 CM004474 OCT79678.1 BC106601 LSYS01009753 OPJ58810.1 HAEE01007246 SBR27266.1 KL321027 KFP54876.1 KL535088 KFO94119.1 HAEH01012150 SBR94063.1 HAED01000872 HAEE01010888 SBQ86717.1 KL216062 KFV67967.1 GGMS01015659 MBY84862.1 CABZ01017629 HAEJ01017589 SBS58046.1 BC083407
Proteomes
UP000005204
UP000218220
UP000283053
UP000037510
UP000007151
UP000053240
+ More
UP000053268 UP000069940 UP000249989 UP000008820 UP000002320 UP000015102 UP000183832 UP000007266 UP000235965 UP000015103 UP000245037 UP000009046 UP000019118 UP000030742 UP000095300 UP000092462 UP000002358 UP000095301 UP000027135 UP000192223 UP000007819 UP000076502 UP000007635 UP000053097 UP000279307 UP000242457 UP000265120 UP000264800 UP000005203 UP000037069 UP000053105 UP000265020 UP000092445 UP000053872 UP000092443 UP000092460 UP000018467 UP000186698 UP000190648 UP000261520 UP000001038 UP000261380 UP000053875 UP000000437 UP000265200
UP000053268 UP000069940 UP000249989 UP000008820 UP000002320 UP000015102 UP000183832 UP000007266 UP000235965 UP000015103 UP000245037 UP000009046 UP000019118 UP000030742 UP000095300 UP000092462 UP000002358 UP000095301 UP000027135 UP000192223 UP000007819 UP000076502 UP000007635 UP000053097 UP000279307 UP000242457 UP000265120 UP000264800 UP000005203 UP000037069 UP000053105 UP000265020 UP000092445 UP000053872 UP000092443 UP000092460 UP000018467 UP000186698 UP000190648 UP000261520 UP000001038 UP000261380 UP000053875 UP000000437 UP000265200
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JFY0
A0A2A4J4F8
A0A3S2LRD3
A0A1E1W4P0
A0A2H1VZ27
A0A0L7LJ51
+ More
A0A212ETP3 S4P8H6 A0A0N1IG38 A0A194PIL3 A0A023EH30 Q17GP9 A0A1Q3FCB8 B0XJI0 A0A0A9WDV8 A0A1L8DCD1 U5EN26 T1GS34 A0A1J1IDS1 D6WVJ5 A0A023EZ69 A0A161N176 A0A2J7QAZ1 T1I8V0 A0A2P8Y2H6 E0VG57 J3JV10 A0A1I8PLW8 A0A0P4VRG2 A0A224XLE5 A0A1B0D5Z2 K7J7U0 A0A1L8EC75 W8BR41 A0A1I8MEV9 A0A2R7VWM3 A0A067QSL4 A0A069DPF0 A0A067QT38 A0A1W4X785 C4WVB2 A0A154PCB6 A0A0A1XME6 A0A2S2NL90 A0A0K8V1N3 G3PE26 A0A026W7E7 A0A2A3E8F0 A0A1B6EKY4 A0A034WXW0 A0A0C9QEM9 A0A3P8UC68 A0A336LJI0 A0A3Q3B7H8 A0A088ABI5 A0A0L0CGD4 A0A0N0U307 A0A1B6LSD0 A0A3Q2FHJ9 C1C2U7 A0A1B0A3F1 A0A3Q2GBJ3 A0A0K8TMF7 A0A091RSK6 A0A2I0M982 A0A1A9YDF9 A0A1B0B009 C1C0D4 A0A2I0M985 A0A3B1K3D5 A0A2H8TU46 A0A1A8ECM6 A0A1L8G708 Q3KPR1 A0A1V4IG26 A0A3B4A2J3 A0A3P8UEL5 A0A1A8K4X6 A0A3B3IN79 A0A091M8P8 A0A091HF74 A0A3B5M4G4 A0A1A8QL70 A0A1A8HRK2 A0A093GLT7 A0A2S2R679 F1QNQ8 A0A1A8VDV5 A0A3P9IWE8 Q5XJ97
A0A212ETP3 S4P8H6 A0A0N1IG38 A0A194PIL3 A0A023EH30 Q17GP9 A0A1Q3FCB8 B0XJI0 A0A0A9WDV8 A0A1L8DCD1 U5EN26 T1GS34 A0A1J1IDS1 D6WVJ5 A0A023EZ69 A0A161N176 A0A2J7QAZ1 T1I8V0 A0A2P8Y2H6 E0VG57 J3JV10 A0A1I8PLW8 A0A0P4VRG2 A0A224XLE5 A0A1B0D5Z2 K7J7U0 A0A1L8EC75 W8BR41 A0A1I8MEV9 A0A2R7VWM3 A0A067QSL4 A0A069DPF0 A0A067QT38 A0A1W4X785 C4WVB2 A0A154PCB6 A0A0A1XME6 A0A2S2NL90 A0A0K8V1N3 G3PE26 A0A026W7E7 A0A2A3E8F0 A0A1B6EKY4 A0A034WXW0 A0A0C9QEM9 A0A3P8UC68 A0A336LJI0 A0A3Q3B7H8 A0A088ABI5 A0A0L0CGD4 A0A0N0U307 A0A1B6LSD0 A0A3Q2FHJ9 C1C2U7 A0A1B0A3F1 A0A3Q2GBJ3 A0A0K8TMF7 A0A091RSK6 A0A2I0M982 A0A1A9YDF9 A0A1B0B009 C1C0D4 A0A2I0M985 A0A3B1K3D5 A0A2H8TU46 A0A1A8ECM6 A0A1L8G708 Q3KPR1 A0A1V4IG26 A0A3B4A2J3 A0A3P8UEL5 A0A1A8K4X6 A0A3B3IN79 A0A091M8P8 A0A091HF74 A0A3B5M4G4 A0A1A8QL70 A0A1A8HRK2 A0A093GLT7 A0A2S2R679 F1QNQ8 A0A1A8VDV5 A0A3P9IWE8 Q5XJ97
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Cytoplasm
Nucleolus
Cytoplasm
Nucleolus
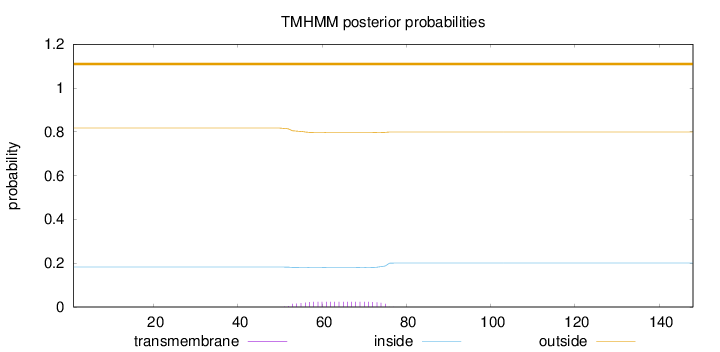
Length:
148
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.50526
Exp number, first 60 AAs:
0.1669
Total prob of N-in:
0.18265
outside
1 - 148
Population Genetic Test Statistics
Pi
112.120632
Theta
124.857032
Tajima's D
-0.313782
CLR
0
CSRT
0.291185440727964
Interpretation
Uncertain
