Gene
KWMTBOMO10780 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008426
Annotation
PREDICTED:_26S_proteasome_non-ATPase_regulatory_subunit_1_[Amyelois_transitella]
Full name
26S proteasome non-ATPase regulatory subunit 1
Alternative Name
26S proteasome regulatory complex subunit p110
26S proteasome regulatory subunit RPN2
26S proteasome regulatory subunit RPN2
Location in the cell
Cytoplasmic Reliability : 2.371
Sequence
CDS
ATGAACATCACGTCTGCCGCGGGGATTATTTCCCTGCTCGATGAGCCCATGTCTGAAGTTAAAGAATTCGCTCTGAAGAGGTTGGACAACATCGTCGATGAGTTTTGGCCAGAAATATCTGAATCTATCGAGAAAATAGAAATCCTGCACGAAGACAAAATTTTCTCCCAGCATCAGCTAGCCGCATTAGTAGCCAGTAAAGTGTACTATCATTTAGGAGCATTTGAAGATTCTTTGACATACGCGCTCGGGGCTGGAGATCTTTTCGACGTGAACGCGAGAAACGAATATGTCGACACTACGATCGCAAAAGCGATCGACTTTTACACTCAAAAGAGAAAGGCTCTCTTTGTGGATAGTTCTGCTGAACTAATTGATCACCGTCTTGAAGACATCGTGAATCGCATGTTCCAGAGGTGCCTTGAAGATGGACAATACAGACAAGCGCTTGGCTTAGCATTGGAGACGCGCCGTATGGATATTTTCGAGGAATCTATTATGAAATCTGATGATATACCAGGAATGCTTCAGTACGCTTTCACAGTAGCAATGAGTCTCCTCCAAAATAAGGGCTTCCGCAGCACTGTACTCCGTTCCTTAGTAACTTTATATAAGGGCCTCAATGTGCCCGATTATGTAAATATGTGCCAGTGTCTTATCTTCTTGGAAGATCCACTAGCTGTAGCAGAGATCCTTGACAAGCTAACACATGGACCACAAGAGTCCGTGTTAATGGCATATCAAATAGCCTTTGATTTGTATGACTCTGCAACTCAACAGTTCTTGGGCAGAGTCTTACAGGCTCTTAGGTGTACTGCACCTATTCCTAGTGCCTTAGGTGGCAAACCACAACCTCAAGGTGGACCTTTCCCAGAATCGTCAATGGAAGTTGACCAACCAGCAACCGAAGAAACCAAAAAGCCAGAGAGAGACATTGACAGTCTAAGTGATGAAGAGAAAGAGCATCAAAAAAGAGTTGAAAAATTGATTTCCATATTAGGAGGAGATGTATCTATAGGCCTCCAACTTCAGTTCTTAATCAGATCTAACCATGCCGATATGCTCATCTTAAAGAACACCAAAGATGCTATAAGGGTATCTATCTGCCACACGGCGACAGTAATAGCCAATGCGTTTATGCATGCAGGTACAACCAGTGATCAGTTTCTTAGGGACAACTTGGAGTGGCTTGCGAGAGCTACAAATTGGGCTAAGTTAACTGTTACAGCTTCTCTAGGAGTAATACATAGGGGTCACGAGAATGAATCCTTAGCTCTCATGCAATCATATCTGCCAAAAGAGGCAGGGCCATCTTCTGGTTATTCAGAAGGAGGTGGTCTCTATGCCTTAGGTTTGATCCATGCTAACCATGGAGCAAACATTATTGATTATCTTTTAACTCAGCTTAAAGATGCACAGAATGAGATGGTAAGACATGGAGGCTGCCTGGGTCTGGGCTTAGCTGCCATGGGCACACATAGACAGGATGTTTATGAACAGCTCAAGTTTAATCTGTATCAAGATGATGCTGTCACAGGAGAAGCAGCAGGTATTGCTATGGGTATGGTCATGCTAGGTTCTAGACATGCTGCTGCCATAGAAGATATGGTTGCCTATGCACAAGAAACACAGCATGAAAAGATCTTGAGAGGCCTTGCTGTTGGCATTGCTTTTACCATGTATGGAAGATTAGAAGAGGCTGATGCTCTGGTTCAGCAACTATTGAGAGACAAAGATCCATTATTGCGACGTGCTGGTTGCTACACTATAGCAACTGCTTATTGTGGCACTGGCAACAATGATTCCATCAGGACTTTGCTTCATGTAGCAGTATCAGATGTTAATGATGATGTTCGTAGGGCAGCTGTGACAGCTTTAGGATTTTTGTTATTTAGAACCCCAGAACAATGTCCCTCAGTAGTGTCGTTGCTGGCTGAATCGTACAATCCTCATGTTCGTTACGGCGCCGCCATGGCTCTGGGTATTGCCTGCGCTGGTACTGGGAATCGTGATGCCATTGGACTCTTGGAACCAATGGTTAAATTCGATCCTGTTAACTTTGTGAGGCAAGGCGCACTAATTGCTTCAGCCATGATTCTCATACAGCAAACCGAAGCGTTATGTCCGAAAGTAACTTATTTCCGTCAACTGTACGCACAAGTTATTTCAAACAAACACGAAGATGTTATGGCGAAGTTCGGAGCAATTTTAGCTCAAGGTATTATCGACGCTGGCGGCCGTAATGTTACCGTTTCACTCCAAAATAGAACTGGGCACATGAATATGCTTGCAGTCGTTGGTATGCTGGTATTTACCCAGTACTGGTATTGGTTCCCACTTGCACACTGCTTATCGCTTGCCTTTACACCAACGTGTATTATTTCACTTAACTCCGATCTTAAAATGCCTCAATTGGAACTGAAGTCCAACTCTAAGCCCTCGCTTTACGCGTATCCAGCTCCGTTGGAAGAAAAGAAACGTGAGGAAAGAGAGAGGGTTACCACTGCTGTGCTAAGTATTGCAGCTGCTAAAGCTCGTAGAAGGGCGCACGGAACTGACGGTTCGGCGAGCAGTGTCACTTCATCTACAACTTCTAAAATGGAAGTTGATGAGGAAGAGAAAAAACCCTCTAAATCCCCTAATATCACAGTTCATAGTAAATCCGATAAAGATGCTCCTAAAGATGCTAAGAAAGACGAAAAAGAAGCAGAGGACAAGGAAGCTAAAGAAAAGAAAGAACCAGAACCGACGTTTGAAATCTTGAGTAATCCTGCCAGAGTGATGAGACAGCAATTGAAGAATTTAACTATTGTCGAGGGTTCAGGTTTCACCCCTTTAAAGGATATCACTATTGGAGGTATTGTCATGTTGAACCACACCGGTGAAGGAGAACAAGTCCTGGTTGAGCCGGTTGCCGCATTCGGCCCGAAGGCAGAGGAAGAGAAGGAGCCTGAACCCCCAGAGCCTTTTGAATATCTGGATGATTGA
Protein
MNITSAAGIISLLDEPMSEVKEFALKRLDNIVDEFWPEISESIEKIEILHEDKIFSQHQLAALVASKVYYHLGAFEDSLTYALGAGDLFDVNARNEYVDTTIAKAIDFYTQKRKALFVDSSAELIDHRLEDIVNRMFQRCLEDGQYRQALGLALETRRMDIFEESIMKSDDIPGMLQYAFTVAMSLLQNKGFRSTVLRSLVTLYKGLNVPDYVNMCQCLIFLEDPLAVAEILDKLTHGPQESVLMAYQIAFDLYDSATQQFLGRVLQALRCTAPIPSALGGKPQPQGGPFPESSMEVDQPATEETKKPERDIDSLSDEEKEHQKRVEKLISILGGDVSIGLQLQFLIRSNHADMLILKNTKDAIRVSICHTATVIANAFMHAGTTSDQFLRDNLEWLARATNWAKLTVTASLGVIHRGHENESLALMQSYLPKEAGPSSGYSEGGGLYALGLIHANHGANIIDYLLTQLKDAQNEMVRHGGCLGLGLAAMGTHRQDVYEQLKFNLYQDDAVTGEAAGIAMGMVMLGSRHAAAIEDMVAYAQETQHEKILRGLAVGIAFTMYGRLEEADALVQQLLRDKDPLLRRAGCYTIATAYCGTGNNDSIRTLLHVAVSDVNDDVRRAAVTALGFLLFRTPEQCPSVVSLLAESYNPHVRYGAAMALGIACAGTGNRDAIGLLEPMVKFDPVNFVRQGALIASAMILIQQTEALCPKVTYFRQLYAQVISNKHEDVMAKFGAILAQGIIDAGGRNVTVSLQNRTGHMNMLAVVGMLVFTQYWYWFPLAHCLSLAFTPTCIISLNSDLKMPQLELKSNSKPSLYAYPAPLEEKKREERERVTTAVLSIAAAKARRRAHGTDGSASSVTSSTTSKMEVDEEEKKPSKSPNITVHSKSDKDAPKDAKKDEKEAEDKEAKEKKEPEPTFEILSNPARVMRQQLKNLTIVEGSGFTPLKDITIGGIVMLNHTGEGEQVLVEPVAAFGPKAEEEKEPEPPEPFEYLDD
Summary
Description
Component of the 26S proteasome, a multiprotein complex involved in the ATP-dependent degradation of ubiquitinated proteins. This complex plays a key role in the maintenance of protein homeostasis by removing misfolded or damaged proteins, which could impair cellular functions, and by removing proteins whose functions are no longer required. Therefore, the proteasome participates in numerous cellular processes, including cell cycle progression, apoptosis, or DNA damage repair.
Acts as a regulatory subunit of the 26S proteasome which is involved in the ATP-dependent degradation of ubiquitinated proteins.
Acts as a regulatory subunit of the 26S proteasome which is involved in the ATP-dependent degradation of ubiquitinated proteins.
Subunit
Component of the 19S proteasome regulatory particle complex. The 26S proteasome consists of a 20S core particle (CP) and two 19S regulatory subunits (RP).
Similarity
Belongs to the proteasome subunit S1 family.
Keywords
Alternative splicing
Complete proteome
Direct protein sequencing
Phosphoprotein
Proteasome
Reference proteome
Repeat
Feature
chain 26S proteasome non-ATPase regulatory subunit 1
splice variant In isoform B.
splice variant In isoform B.
Uniprot
A0A2A4J4L9
A0A2H1W0Q6
A0A212ETL7
A0A3S2TDE2
A0A0N1IHD5
A0A194PCC3
+ More
A0A1Y1LWY3 A0A182GGV4 A0A182GCZ1 A0A182QI93 Q17GS7 A0A1S4F345 U5EZB8 A0A182RPD3 A0A182P2L9 A0A182M3M5 A0A182VRT0 A0A182IQ81 A0A1Q3F7S0 A0A182WSV8 B0WHS6 A0A1L8DSW1 W5J7R6 A0A1L8DTE1 A0A2M4A606 A0A182FNK1 A0A2M4BBI5 A0A2M4BBK6 A0A0C9QZ65 A0A336MJX1 A0A1A9Y8Y9 A0A1A9V941 A0A1B0BGT1 A0A1B0G973 A0A1B0AGP8 A0A3L8DKX1 A0A158NMH3 D3TNJ0 A0A151JW54 A0A1A9WTQ9 A0A1I8NXP0 N6TU34 A0A1Q3F7I5 A0A195AZT6 A0A195CW03 B4K5W8 A0A0L7LJU1 T1PAM5 A0A1W4UIP4 B4NFM4 B4PQ60 B4HZ72 A0A084W312 A0A0B4KHE9 Q9V3P6 A0A0B4KI10 A0A182XUZ7 B4MBT7 A0A182N7C7 B3P5K1 A0A151WUT8 A0A182HLR2 A0A182VH90 A0A1B6C4U5 A0A182TRH7 A0A026WFH5 Q7PY49 A0A182JU48 A0A2M4A5L8 A0A2P8XR50 W8AWT3 A0A034VUA6 A0A232FBX0 A0A0K8V7J3 K7IPB1 A0A0A1WQ05 A0A0L0CEV3 A0A1B6GG98 B3M1I7 A0A0M3QY66 B4JYI2 A0A3B0K6D6 A0A2L2XVN8 A0A182LA62 B5DYD3 A0A069DZN6 A0A224XJ34 A0A0P4VTW8 A0A023F4B3 E0VDU8 A0A087SVX7 A0A2S2Q2S8 A0A2H8TNJ6 J9JRM2 A0A0A9WRY6 A0A0K8S9P2 A0A293MXK4 A0A1Z5KUL7 A0A0V0G415 A0A0P5RGH1
A0A1Y1LWY3 A0A182GGV4 A0A182GCZ1 A0A182QI93 Q17GS7 A0A1S4F345 U5EZB8 A0A182RPD3 A0A182P2L9 A0A182M3M5 A0A182VRT0 A0A182IQ81 A0A1Q3F7S0 A0A182WSV8 B0WHS6 A0A1L8DSW1 W5J7R6 A0A1L8DTE1 A0A2M4A606 A0A182FNK1 A0A2M4BBI5 A0A2M4BBK6 A0A0C9QZ65 A0A336MJX1 A0A1A9Y8Y9 A0A1A9V941 A0A1B0BGT1 A0A1B0G973 A0A1B0AGP8 A0A3L8DKX1 A0A158NMH3 D3TNJ0 A0A151JW54 A0A1A9WTQ9 A0A1I8NXP0 N6TU34 A0A1Q3F7I5 A0A195AZT6 A0A195CW03 B4K5W8 A0A0L7LJU1 T1PAM5 A0A1W4UIP4 B4NFM4 B4PQ60 B4HZ72 A0A084W312 A0A0B4KHE9 Q9V3P6 A0A0B4KI10 A0A182XUZ7 B4MBT7 A0A182N7C7 B3P5K1 A0A151WUT8 A0A182HLR2 A0A182VH90 A0A1B6C4U5 A0A182TRH7 A0A026WFH5 Q7PY49 A0A182JU48 A0A2M4A5L8 A0A2P8XR50 W8AWT3 A0A034VUA6 A0A232FBX0 A0A0K8V7J3 K7IPB1 A0A0A1WQ05 A0A0L0CEV3 A0A1B6GG98 B3M1I7 A0A0M3QY66 B4JYI2 A0A3B0K6D6 A0A2L2XVN8 A0A182LA62 B5DYD3 A0A069DZN6 A0A224XJ34 A0A0P4VTW8 A0A023F4B3 E0VDU8 A0A087SVX7 A0A2S2Q2S8 A0A2H8TNJ6 J9JRM2 A0A0A9WRY6 A0A0K8S9P2 A0A293MXK4 A0A1Z5KUL7 A0A0V0G415 A0A0P5RGH1
Pubmed
22118469
26354079
28004739
26483478
17510324
20920257
+ More
23761445 30249741 21347285 20353571 23537049 17994087 26227816 25315136 17550304 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10893261 11157774 12537569 18327897 25244985 24508170 12364791 14747013 17210077 29403074 24495485 25348373 28648823 20075255 25830018 26108605 26561354 20966253 15632085 26334808 27129103 25474469 20566863 25401762 26823975 28528879
23761445 30249741 21347285 20353571 23537049 17994087 26227816 25315136 17550304 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 10893261 11157774 12537569 18327897 25244985 24508170 12364791 14747013 17210077 29403074 24495485 25348373 28648823 20075255 25830018 26108605 26561354 20966253 15632085 26334808 27129103 25474469 20566863 25401762 26823975 28528879
EMBL
NWSH01003288
PCG66618.1
ODYU01005346
SOQ46094.1
AGBW02012556
OWR44836.1
+ More
RSAL01000298 RVE42794.1 KQ460556 KPJ13999.1 KQ459606 KPI90931.1 GEZM01044900 JAV78053.1 JXUM01062546 KQ562204 KXJ76426.1 JXUM01160558 KQ573741 KXJ67908.1 AXCN02000143 CH477256 EAT45860.1 GANO01001077 JAB58794.1 AXCM01001440 GFDL01011476 JAV23569.1 DS231939 EDS27943.1 GFDF01004528 JAV09556.1 ADMH02002001 ETN60021.1 GFDF01004529 JAV09555.1 GGFK01002737 MBW36058.1 GGFJ01001273 MBW50414.1 GGFJ01001272 MBW50413.1 GBYB01005987 JAG75754.1 UFQT01000671 SSX26358.1 JXJN01013985 CCAG010000643 QOIP01000007 RLU20559.1 ADTU01020488 ADTU01020489 ADTU01020490 ADTU01020491 ADTU01020492 ADTU01020493 EZ422992 ADD19268.1 KQ981676 KYN38150.1 APGK01018904 KB740085 KB632303 ENN81538.1 ERL91804.1 GFDL01011519 JAV23526.1 KQ976697 KYM77545.1 KQ977231 KYN04717.1 CH933806 EDW16205.1 JTDY01000907 KOB75476.1 KA645787 AFP60416.1 CH964251 EDW83091.2 CM000160 EDW98329.2 CH480819 EDW53329.1 ATLV01019778 KE525279 KFB44606.1 AE014297 AGB96438.1 AF145303 AF186594 AF312231 AY069503 BT001598 BT044102 AGB96437.1 CH940656 EDW58558.2 CH954182 EDV53251.1 KQ982727 KYQ51606.1 APCN01000911 GEDC01028979 JAS08319.1 KK107238 EZA54812.1 AAAB01008987 EAA01256.5 GGFK01002720 MBW36041.1 PYGN01001493 PSN34480.1 GAMC01015998 JAB90557.1 GAKP01013290 JAC45662.1 NNAY01000447 OXU28321.1 GDHF01017528 JAI34786.1 GBXI01013375 JAD00917.1 JRES01000501 KNC30747.1 GECZ01008436 JAS61333.1 CH902617 EDV43278.2 CP012526 ALC47143.1 CH916377 EDV90744.1 OUUW01000005 SPP81186.1 IAAA01005931 LAA00171.1 CM000070 EDY67585.2 GBGD01000325 JAC88564.1 GFTR01008385 JAW08041.1 GDKW01000506 JAI56089.1 GBBI01002366 JAC16346.1 DS235088 EEB11554.1 KK112194 KFM57016.1 GGMS01002820 MBY72023.1 GFXV01003938 MBW15743.1 ABLF02016086 GBHO01032387 GDHC01006042 JAG11217.1 JAQ12587.1 GBRD01015932 JAG49894.1 GFWV01020815 MAA45543.1 GFJQ02008164 JAV98805.1 GECL01003700 JAP02424.1 GDIQ01120373 JAL31353.1
RSAL01000298 RVE42794.1 KQ460556 KPJ13999.1 KQ459606 KPI90931.1 GEZM01044900 JAV78053.1 JXUM01062546 KQ562204 KXJ76426.1 JXUM01160558 KQ573741 KXJ67908.1 AXCN02000143 CH477256 EAT45860.1 GANO01001077 JAB58794.1 AXCM01001440 GFDL01011476 JAV23569.1 DS231939 EDS27943.1 GFDF01004528 JAV09556.1 ADMH02002001 ETN60021.1 GFDF01004529 JAV09555.1 GGFK01002737 MBW36058.1 GGFJ01001273 MBW50414.1 GGFJ01001272 MBW50413.1 GBYB01005987 JAG75754.1 UFQT01000671 SSX26358.1 JXJN01013985 CCAG010000643 QOIP01000007 RLU20559.1 ADTU01020488 ADTU01020489 ADTU01020490 ADTU01020491 ADTU01020492 ADTU01020493 EZ422992 ADD19268.1 KQ981676 KYN38150.1 APGK01018904 KB740085 KB632303 ENN81538.1 ERL91804.1 GFDL01011519 JAV23526.1 KQ976697 KYM77545.1 KQ977231 KYN04717.1 CH933806 EDW16205.1 JTDY01000907 KOB75476.1 KA645787 AFP60416.1 CH964251 EDW83091.2 CM000160 EDW98329.2 CH480819 EDW53329.1 ATLV01019778 KE525279 KFB44606.1 AE014297 AGB96438.1 AF145303 AF186594 AF312231 AY069503 BT001598 BT044102 AGB96437.1 CH940656 EDW58558.2 CH954182 EDV53251.1 KQ982727 KYQ51606.1 APCN01000911 GEDC01028979 JAS08319.1 KK107238 EZA54812.1 AAAB01008987 EAA01256.5 GGFK01002720 MBW36041.1 PYGN01001493 PSN34480.1 GAMC01015998 JAB90557.1 GAKP01013290 JAC45662.1 NNAY01000447 OXU28321.1 GDHF01017528 JAI34786.1 GBXI01013375 JAD00917.1 JRES01000501 KNC30747.1 GECZ01008436 JAS61333.1 CH902617 EDV43278.2 CP012526 ALC47143.1 CH916377 EDV90744.1 OUUW01000005 SPP81186.1 IAAA01005931 LAA00171.1 CM000070 EDY67585.2 GBGD01000325 JAC88564.1 GFTR01008385 JAW08041.1 GDKW01000506 JAI56089.1 GBBI01002366 JAC16346.1 DS235088 EEB11554.1 KK112194 KFM57016.1 GGMS01002820 MBY72023.1 GFXV01003938 MBW15743.1 ABLF02016086 GBHO01032387 GDHC01006042 JAG11217.1 JAQ12587.1 GBRD01015932 JAG49894.1 GFWV01020815 MAA45543.1 GFJQ02008164 JAV98805.1 GECL01003700 JAP02424.1 GDIQ01120373 JAL31353.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
UP000069940
+ More
UP000249989 UP000075886 UP000008820 UP000075900 UP000075885 UP000075883 UP000075920 UP000075880 UP000076407 UP000002320 UP000000673 UP000069272 UP000092443 UP000078200 UP000092460 UP000092444 UP000092445 UP000279307 UP000005205 UP000078541 UP000091820 UP000095300 UP000019118 UP000030742 UP000078540 UP000078542 UP000009192 UP000037510 UP000095301 UP000192221 UP000007798 UP000002282 UP000001292 UP000030765 UP000000803 UP000076408 UP000008792 UP000075884 UP000008711 UP000075809 UP000075840 UP000075903 UP000075902 UP000053097 UP000007062 UP000075881 UP000245037 UP000215335 UP000002358 UP000037069 UP000007801 UP000092553 UP000001070 UP000268350 UP000075882 UP000001819 UP000009046 UP000054359 UP000007819
UP000249989 UP000075886 UP000008820 UP000075900 UP000075885 UP000075883 UP000075920 UP000075880 UP000076407 UP000002320 UP000000673 UP000069272 UP000092443 UP000078200 UP000092460 UP000092444 UP000092445 UP000279307 UP000005205 UP000078541 UP000091820 UP000095300 UP000019118 UP000030742 UP000078540 UP000078542 UP000009192 UP000037510 UP000095301 UP000192221 UP000007798 UP000002282 UP000001292 UP000030765 UP000000803 UP000076408 UP000008792 UP000075884 UP000008711 UP000075809 UP000075840 UP000075903 UP000075902 UP000053097 UP000007062 UP000075881 UP000245037 UP000215335 UP000002358 UP000037069 UP000007801 UP000092553 UP000001070 UP000268350 UP000075882 UP000001819 UP000009046 UP000054359 UP000007819
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2A4J4L9
A0A2H1W0Q6
A0A212ETL7
A0A3S2TDE2
A0A0N1IHD5
A0A194PCC3
+ More
A0A1Y1LWY3 A0A182GGV4 A0A182GCZ1 A0A182QI93 Q17GS7 A0A1S4F345 U5EZB8 A0A182RPD3 A0A182P2L9 A0A182M3M5 A0A182VRT0 A0A182IQ81 A0A1Q3F7S0 A0A182WSV8 B0WHS6 A0A1L8DSW1 W5J7R6 A0A1L8DTE1 A0A2M4A606 A0A182FNK1 A0A2M4BBI5 A0A2M4BBK6 A0A0C9QZ65 A0A336MJX1 A0A1A9Y8Y9 A0A1A9V941 A0A1B0BGT1 A0A1B0G973 A0A1B0AGP8 A0A3L8DKX1 A0A158NMH3 D3TNJ0 A0A151JW54 A0A1A9WTQ9 A0A1I8NXP0 N6TU34 A0A1Q3F7I5 A0A195AZT6 A0A195CW03 B4K5W8 A0A0L7LJU1 T1PAM5 A0A1W4UIP4 B4NFM4 B4PQ60 B4HZ72 A0A084W312 A0A0B4KHE9 Q9V3P6 A0A0B4KI10 A0A182XUZ7 B4MBT7 A0A182N7C7 B3P5K1 A0A151WUT8 A0A182HLR2 A0A182VH90 A0A1B6C4U5 A0A182TRH7 A0A026WFH5 Q7PY49 A0A182JU48 A0A2M4A5L8 A0A2P8XR50 W8AWT3 A0A034VUA6 A0A232FBX0 A0A0K8V7J3 K7IPB1 A0A0A1WQ05 A0A0L0CEV3 A0A1B6GG98 B3M1I7 A0A0M3QY66 B4JYI2 A0A3B0K6D6 A0A2L2XVN8 A0A182LA62 B5DYD3 A0A069DZN6 A0A224XJ34 A0A0P4VTW8 A0A023F4B3 E0VDU8 A0A087SVX7 A0A2S2Q2S8 A0A2H8TNJ6 J9JRM2 A0A0A9WRY6 A0A0K8S9P2 A0A293MXK4 A0A1Z5KUL7 A0A0V0G415 A0A0P5RGH1
A0A1Y1LWY3 A0A182GGV4 A0A182GCZ1 A0A182QI93 Q17GS7 A0A1S4F345 U5EZB8 A0A182RPD3 A0A182P2L9 A0A182M3M5 A0A182VRT0 A0A182IQ81 A0A1Q3F7S0 A0A182WSV8 B0WHS6 A0A1L8DSW1 W5J7R6 A0A1L8DTE1 A0A2M4A606 A0A182FNK1 A0A2M4BBI5 A0A2M4BBK6 A0A0C9QZ65 A0A336MJX1 A0A1A9Y8Y9 A0A1A9V941 A0A1B0BGT1 A0A1B0G973 A0A1B0AGP8 A0A3L8DKX1 A0A158NMH3 D3TNJ0 A0A151JW54 A0A1A9WTQ9 A0A1I8NXP0 N6TU34 A0A1Q3F7I5 A0A195AZT6 A0A195CW03 B4K5W8 A0A0L7LJU1 T1PAM5 A0A1W4UIP4 B4NFM4 B4PQ60 B4HZ72 A0A084W312 A0A0B4KHE9 Q9V3P6 A0A0B4KI10 A0A182XUZ7 B4MBT7 A0A182N7C7 B3P5K1 A0A151WUT8 A0A182HLR2 A0A182VH90 A0A1B6C4U5 A0A182TRH7 A0A026WFH5 Q7PY49 A0A182JU48 A0A2M4A5L8 A0A2P8XR50 W8AWT3 A0A034VUA6 A0A232FBX0 A0A0K8V7J3 K7IPB1 A0A0A1WQ05 A0A0L0CEV3 A0A1B6GG98 B3M1I7 A0A0M3QY66 B4JYI2 A0A3B0K6D6 A0A2L2XVN8 A0A182LA62 B5DYD3 A0A069DZN6 A0A224XJ34 A0A0P4VTW8 A0A023F4B3 E0VDU8 A0A087SVX7 A0A2S2Q2S8 A0A2H8TNJ6 J9JRM2 A0A0A9WRY6 A0A0K8S9P2 A0A293MXK4 A0A1Z5KUL7 A0A0V0G415 A0A0P5RGH1
PDB
6MSK
E-value=0,
Score=2961
Ontologies
GO
PANTHER
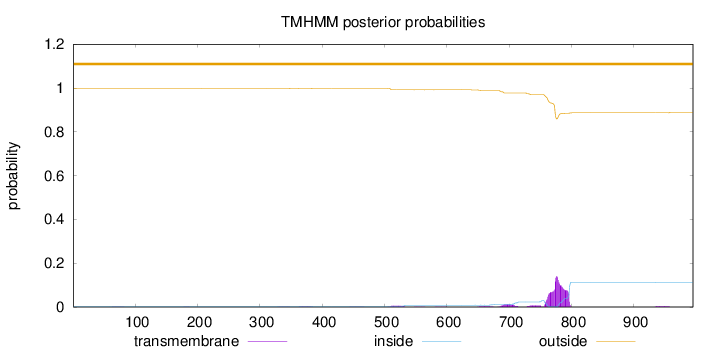
Topology
Length:
995
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.88494
Exp number, first 60 AAs:
0.00028
Total prob of N-in:
0.00314
outside
1 - 995
Population Genetic Test Statistics
Pi
251.05917
Theta
233.843926
Tajima's D
0.244253
CLR
0.044104
CSRT
0.437778111094445
Interpretation
Uncertain
