Pre Gene Modal
BGIBMGA008392
Annotation
signal_recognition_particle_receptor_alpha_subunit_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.898 Mitochondrial Reliability : 1.323
Sequence
CDS
ATGTTAGATTTATTCAGTATTTTCAGTAAAGGGGGTATTGTGTTATGGTGTTTCCAAAGTACTAGTGAAATATTTTCGCCATCCGTGAACGCTTTAATACGTAGTGTAATATTGCAAGAACGCTCTGGCAATAACACATTCAATCATAATGCCTTAACACTACAATACAAACTAGATAATGAGTTTGAACTGGTTTTTGTAGTTGCATACCAACGCATACTCCAGTTATCATATGTGGATAAGTTCCTCAATGATGTCCACTTGGAGTTCAGGGACAAGTACAAGAATGAGCTCCAGACAGGTCCCTGTATCGTTGATTTTAATTTTAAAACTACCTTTGATAAAGTGCTGAAGGAATGTGAAATATGGGCAAAGTCACAAGCTAAAATTCCAAAACAGATGAGGACTTTTGAGGATTCCCAAAAATCCAAAAAGACGGTTGCATCTATGATCGAACGCAAAGGAGATGAGAAAGGGAAGAAAACTGTCAAAATAGTCGAAAGTAAAAATCTTACAAATGGTAAAGCAGAGGAAGTTACAAATAATACATATGATGATGATGTCATCGCTGCAAACAGAGCCAAGCTAGCACAGAAGCTGGCATCGAAAACAAAGAAGGAACCCAAGAAATCTCCAAAGAGTCCCCAGAAGGCAGACCGCGTGAAGCAGCCCCGCGTGTGGGAGCTCGGCGGCGACACGCGCGACGTCGCTGTGCTGGACTACAGTACTGACCAACCAGGAGATGCTGCTACCTTACTGCCGGACACGCAGTTGATTGGTCAAATGACTGGTGCCATTCGTGACTTGGAAGTAGAATCCGAGGAGAGCAGCAGTGAGGAAGAGGAGGTAACTTCTCAGCCGACTTCCAAGAAAACTGGAGGAATGTTCAGTTTATTAAAAGGTCTAGTCGGTGCAAAAGCATTGTCAGAGGAAACGATGCGACCGGTGTTGGACAAGCTTCGTGATCACCTCATTGGGAAGAACGTCGCCGCGGACATCGCCAATAAGCTCTGTGATAGTGTTGCCATCAAACTAGAGGGGAAGGTGCTAGGTACCTTCGACAGTGTTGCCAAGACGGTGAAGGCTACCCTCACCGAGTCCCTCGTGCGGATACTGTCGCCGAAGCGTCGCGTGGACATCCTGCGGGACTGTCTGCACGCCAAGCAGGAGGGCAGGCCCTACGTCATGGCGTTCTGCGGGGTGAACGGCGTCGGTAAATCGACGAACCTGGCGAAGATCTGCTTCTGGTTGATCGAGAACGATCTGTCGGTGCTGATAGCGGCGTGCGACACGTTCCGCGCCGGCGCCGTGGAGCAGCTGCGCACGCACACGCGACACCTCAACGCGCTGCACCCGCCCGCGCGCCACCACGCACGACACATGGTCACGCTCTACGAGAAGGGATACGGCAAAGACGCGGCCGGCATAGCAATGGAAGCAATACGATACGCGTCTGACACTAAAACCGACGTCGTGCTCATCGACACTGCGGGCCGAATGCAGGACAACGAGCCTCTGATGCGGGCCCTCGCCAAGCTCATCGCGGTCAACGAACCCGACACGGTGCTCTTCGTGGGCGAGGCTCTCGTAGGGAACGAGGCCGTCGACCAGCTGGTCAAGTTCAACCAGGCCCTCGCCGACTACAGCTCGTCCAGCAGTCCGCACGTCATCGACGGAATAGTGCTCACCAAGTTCGATACTATCGACGATAAAGTCGGCGCCGCCATATCGATGACGTACATCACCGGGCAACCGATAGTGTTCGTCGGAACGGGACAAACGTACACCGATTTGAAAGCGCTGAACGCCAACGCTGTAGTTCACGCCTTGATGAAGTGA
Protein
MLDLFSIFSKGGIVLWCFQSTSEIFSPSVNALIRSVILQERSGNNTFNHNALTLQYKLDNEFELVFVVAYQRILQLSYVDKFLNDVHLEFRDKYKNELQTGPCIVDFNFKTTFDKVLKECEIWAKSQAKIPKQMRTFEDSQKSKKTVASMIERKGDEKGKKTVKIVESKNLTNGKAEEVTNNTYDDDVIAANRAKLAQKLASKTKKEPKKSPKSPQKADRVKQPRVWELGGDTRDVAVLDYSTDQPGDAATLLPDTQLIGQMTGAIRDLEVESEESSSEEEEVTSQPTSKKTGGMFSLLKGLVGAKALSEETMRPVLDKLRDHLIGKNVAADIANKLCDSVAIKLEGKVLGTFDSVAKTVKATLTESLVRILSPKRRVDILRDCLHAKQEGRPYVMAFCGVNGVGKSTNLAKICFWLIENDLSVLIAACDTFRAGAVEQLRTHTRHLNALHPPARHHARHMVTLYEKGYGKDAAGIAMEAIRYASDTKTDVVLIDTAGRMQDNEPLMRALAKLIAVNEPDTVLFVGEALVGNEAVDQLVKFNQALADYSSSSSPHVIDGIVLTKFDTIDDKVGAAISMTYITGQPIVFVGTGQTYTDLKALNANAVVHALMK
Summary
Uniprot
Q0N2R5
H9JFU5
A0A2A4J427
A0A2H1VZ60
A0A194PCK6
A0A212ETK6
+ More
A0A0N1IPE4 K7J9K8 A0A069DVD0 A0A023F8T3 A0A088AP45 A0A195CAB7 F4X412 A0A151X6E4 V9IGE9 A0A195BGZ4 A0A195FPB6 A0A1Y1LJQ3 A0A154PNW3 U5EYE4 A0A067QJ19 E2B3Y8 A0A0M8ZVN2 E9JC37 A0A2A3EQB1 A0A1L8DVM6 A0A0A9Y262 A0A310SDV2 A0A1B6F339 A0A1B0CC27 A0A026WD67 E2ARX2 A0A158NJL0 A0A0P4VUW5 R4FNW4 Q178J5 A0A084VFJ0 A0A1B6IS61 D2A3J6 A0A023EUH7 A0A1J1J681 A0A182KG01 A0A182WYD0 A0A182U4T0 Q7PSJ1 A0A182KVP4 A0A182I5F8 A0A182YRK4 A0A182V256 A0A182PJD2 A0A182GMC6 A0A182NC68 A0A182IZY4 A0A2M4A4A8 A0A182RW26 B0WDS3 A0A2M4A4A5 A0A182QWA7 W5JGQ4 A0A1Q3FHJ0 A0A1Q3FHM5 A0A0K8TMF5 A0A0L7QSX7 A0A2M4BH03 A0A336MT09 A0A2M4BGY1 A0A2M4BGY6 J3JTJ5 A0A336MV84 A0A182F467 A0A2M3Z0M9 A0A182M497 A0A1W4XHC4 A0A1S3DAA0 A0A2S2QQE9 A0A0C9PLB3 A0A2S2P6A5 A0A2H8U1X4 J9K9M6 A0A2R7WSB6 A0A1Z5KVX9 E0VIM1 A0A162D7B5 A0A2R5LN40 A0A0P5XM93 A0A0P4Y2R0 E9G6J2 A0A0P5A1J5 A0A0P4XZY5 A0A0P5A2K5 A0A293M1B2 A0A0K8RIY4 A0A0P6E389 A0A0P5ZH84 A0A2L2Y3W6 A0A0P5MS51 A0A087UZW9 A0A023GD26 A0A1S3JUS1 A0A131XEV6
A0A0N1IPE4 K7J9K8 A0A069DVD0 A0A023F8T3 A0A088AP45 A0A195CAB7 F4X412 A0A151X6E4 V9IGE9 A0A195BGZ4 A0A195FPB6 A0A1Y1LJQ3 A0A154PNW3 U5EYE4 A0A067QJ19 E2B3Y8 A0A0M8ZVN2 E9JC37 A0A2A3EQB1 A0A1L8DVM6 A0A0A9Y262 A0A310SDV2 A0A1B6F339 A0A1B0CC27 A0A026WD67 E2ARX2 A0A158NJL0 A0A0P4VUW5 R4FNW4 Q178J5 A0A084VFJ0 A0A1B6IS61 D2A3J6 A0A023EUH7 A0A1J1J681 A0A182KG01 A0A182WYD0 A0A182U4T0 Q7PSJ1 A0A182KVP4 A0A182I5F8 A0A182YRK4 A0A182V256 A0A182PJD2 A0A182GMC6 A0A182NC68 A0A182IZY4 A0A2M4A4A8 A0A182RW26 B0WDS3 A0A2M4A4A5 A0A182QWA7 W5JGQ4 A0A1Q3FHJ0 A0A1Q3FHM5 A0A0K8TMF5 A0A0L7QSX7 A0A2M4BH03 A0A336MT09 A0A2M4BGY1 A0A2M4BGY6 J3JTJ5 A0A336MV84 A0A182F467 A0A2M3Z0M9 A0A182M497 A0A1W4XHC4 A0A1S3DAA0 A0A2S2QQE9 A0A0C9PLB3 A0A2S2P6A5 A0A2H8U1X4 J9K9M6 A0A2R7WSB6 A0A1Z5KVX9 E0VIM1 A0A162D7B5 A0A2R5LN40 A0A0P5XM93 A0A0P4Y2R0 E9G6J2 A0A0P5A1J5 A0A0P4XZY5 A0A0P5A2K5 A0A293M1B2 A0A0K8RIY4 A0A0P6E389 A0A0P5ZH84 A0A2L2Y3W6 A0A0P5MS51 A0A087UZW9 A0A023GD26 A0A1S3JUS1 A0A131XEV6
Pubmed
19121390
26354079
22118469
20075255
26334808
25474469
+ More
21719571 28004739 24845553 20798317 21282665 25401762 26823975 24508170 30249741 21347285 27129103 17510324 24438588 18362917 19820115 24945155 12364791 14747013 17210077 20966253 25244985 26483478 20920257 23761445 26369729 22516182 23537049 28528879 20566863 21292972 26561354 28049606
21719571 28004739 24845553 20798317 21282665 25401762 26823975 24508170 30249741 21347285 27129103 17510324 24438588 18362917 19820115 24945155 12364791 14747013 17210077 20966253 25244985 26483478 20920257 23761445 26369729 22516182 23537049 28528879 20566863 21292972 26561354 28049606
EMBL
DQ646416
ABH10806.1
BABH01001596
BABH01001597
NWSH01003288
PCG66619.1
+ More
ODYU01005346 SOQ46093.1 KQ459606 KPI90932.1 AGBW02012556 OWR44835.1 KQ460556 KPJ13998.1 AAZX01006880 GBGD01000846 JAC88043.1 GBBI01000876 JAC17836.1 KQ978081 KYM97131.1 GL888624 EGI58945.1 KQ982482 KYQ55973.1 JR041334 AEY59414.1 KQ976490 KYM83439.1 KQ981424 KYN41789.1 GEZM01057672 JAV72165.1 KQ435007 KZC13571.1 GANO01000349 JAB59522.1 KK853289 KDR08912.1 GL445421 EFN89594.1 KQ435859 KOX70543.1 GL771706 EFZ09612.1 KZ288193 PBC33983.1 GFDF01003739 JAV10345.1 GBHO01018396 GBRD01005219 GDHC01021235 JAG25208.1 JAG60602.1 JAP97393.1 KQ762518 OAD55792.1 GECZ01025202 JAS44567.1 AJWK01006097 KK107260 QOIP01000001 EZA54007.1 RLU27192.1 GL442192 EFN63821.1 ADTU01018039 ADTU01018040 ADTU01018041 GDKW01000766 JAI55829.1 ACPB03016850 GAHY01000408 JAA77102.1 CH477362 EAT42647.1 ATLV01012419 KE524791 KFB36734.1 GECU01017934 JAS89772.1 KQ971338 EFA02325.1 GAPW01000538 JAC13060.1 CVRI01000070 CRL07276.1 AAAB01008823 EAA05506.5 APCN01003437 JXUM01074362 KQ562824 KXJ75055.1 GGFK01002260 MBW35581.1 DS231900 EDS44826.1 GGFK01002268 MBW35589.1 AXCN02002119 ADMH02001239 ETN63542.1 GFDL01008072 JAV26973.1 GFDL01008001 JAV27044.1 GDAI01002265 JAI15338.1 KQ414757 KOC61576.1 GGFJ01003176 MBW52317.1 UFQT01002342 SSX33240.1 GGFJ01003174 MBW52315.1 GGFJ01003175 MBW52316.1 BT126552 KB632420 AEE61516.1 ERL95318.1 UFQS01002518 UFQT01002518 SSX14199.1 SSX33615.1 GGFM01001336 MBW22087.1 AXCM01002806 GGMS01010796 MBY79999.1 GBYB01001828 GBYB01011017 JAG71595.1 JAG80784.1 GGMR01012341 MBY24960.1 GFXV01007663 MBW19468.1 ABLF02014861 KK855412 PTY22446.1 GFJQ02007760 JAV99209.1 DS235201 EEB13227.1 LRGB01002384 KZS07914.1 GGLE01006827 MBY10953.1 GDIP01070243 JAM33472.1 GDIP01233380 JAI90021.1 GL732533 EFX84945.1 GDIP01217879 JAJ05523.1 GDIP01234479 JAI88922.1 GDIP01205176 JAJ18226.1 GFWV01009270 MAA33999.1 GADI01003329 JAA70479.1 GDIQ01083203 JAN11534.1 GDIP01043752 JAM59963.1 IAAA01003051 LAA02851.1 GDIQ01174809 JAK76916.1 KK122523 KFM82908.1 GBBM01003651 JAC31767.1 GEFH01003699 JAP64882.1
ODYU01005346 SOQ46093.1 KQ459606 KPI90932.1 AGBW02012556 OWR44835.1 KQ460556 KPJ13998.1 AAZX01006880 GBGD01000846 JAC88043.1 GBBI01000876 JAC17836.1 KQ978081 KYM97131.1 GL888624 EGI58945.1 KQ982482 KYQ55973.1 JR041334 AEY59414.1 KQ976490 KYM83439.1 KQ981424 KYN41789.1 GEZM01057672 JAV72165.1 KQ435007 KZC13571.1 GANO01000349 JAB59522.1 KK853289 KDR08912.1 GL445421 EFN89594.1 KQ435859 KOX70543.1 GL771706 EFZ09612.1 KZ288193 PBC33983.1 GFDF01003739 JAV10345.1 GBHO01018396 GBRD01005219 GDHC01021235 JAG25208.1 JAG60602.1 JAP97393.1 KQ762518 OAD55792.1 GECZ01025202 JAS44567.1 AJWK01006097 KK107260 QOIP01000001 EZA54007.1 RLU27192.1 GL442192 EFN63821.1 ADTU01018039 ADTU01018040 ADTU01018041 GDKW01000766 JAI55829.1 ACPB03016850 GAHY01000408 JAA77102.1 CH477362 EAT42647.1 ATLV01012419 KE524791 KFB36734.1 GECU01017934 JAS89772.1 KQ971338 EFA02325.1 GAPW01000538 JAC13060.1 CVRI01000070 CRL07276.1 AAAB01008823 EAA05506.5 APCN01003437 JXUM01074362 KQ562824 KXJ75055.1 GGFK01002260 MBW35581.1 DS231900 EDS44826.1 GGFK01002268 MBW35589.1 AXCN02002119 ADMH02001239 ETN63542.1 GFDL01008072 JAV26973.1 GFDL01008001 JAV27044.1 GDAI01002265 JAI15338.1 KQ414757 KOC61576.1 GGFJ01003176 MBW52317.1 UFQT01002342 SSX33240.1 GGFJ01003174 MBW52315.1 GGFJ01003175 MBW52316.1 BT126552 KB632420 AEE61516.1 ERL95318.1 UFQS01002518 UFQT01002518 SSX14199.1 SSX33615.1 GGFM01001336 MBW22087.1 AXCM01002806 GGMS01010796 MBY79999.1 GBYB01001828 GBYB01011017 JAG71595.1 JAG80784.1 GGMR01012341 MBY24960.1 GFXV01007663 MBW19468.1 ABLF02014861 KK855412 PTY22446.1 GFJQ02007760 JAV99209.1 DS235201 EEB13227.1 LRGB01002384 KZS07914.1 GGLE01006827 MBY10953.1 GDIP01070243 JAM33472.1 GDIP01233380 JAI90021.1 GL732533 EFX84945.1 GDIP01217879 JAJ05523.1 GDIP01234479 JAI88922.1 GDIP01205176 JAJ18226.1 GFWV01009270 MAA33999.1 GADI01003329 JAA70479.1 GDIQ01083203 JAN11534.1 GDIP01043752 JAM59963.1 IAAA01003051 LAA02851.1 GDIQ01174809 JAK76916.1 KK122523 KFM82908.1 GBBM01003651 JAC31767.1 GEFH01003699 JAP64882.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000002358
+ More
UP000005203 UP000078542 UP000007755 UP000075809 UP000078540 UP000078541 UP000076502 UP000027135 UP000008237 UP000053105 UP000242457 UP000092461 UP000053097 UP000279307 UP000000311 UP000005205 UP000015103 UP000008820 UP000030765 UP000007266 UP000183832 UP000075881 UP000076407 UP000075902 UP000007062 UP000075882 UP000075840 UP000076408 UP000075903 UP000075885 UP000069940 UP000249989 UP000075884 UP000075880 UP000075900 UP000002320 UP000075886 UP000000673 UP000053825 UP000030742 UP000069272 UP000075883 UP000192223 UP000079169 UP000007819 UP000009046 UP000076858 UP000000305 UP000054359 UP000085678
UP000005203 UP000078542 UP000007755 UP000075809 UP000078540 UP000078541 UP000076502 UP000027135 UP000008237 UP000053105 UP000242457 UP000092461 UP000053097 UP000279307 UP000000311 UP000005205 UP000015103 UP000008820 UP000030765 UP000007266 UP000183832 UP000075881 UP000076407 UP000075902 UP000007062 UP000075882 UP000075840 UP000076408 UP000075903 UP000075885 UP000069940 UP000249989 UP000075884 UP000075880 UP000075900 UP000002320 UP000075886 UP000000673 UP000053825 UP000030742 UP000069272 UP000075883 UP000192223 UP000079169 UP000007819 UP000009046 UP000076858 UP000000305 UP000054359 UP000085678
Interpro
Gene 3D
ProteinModelPortal
Q0N2R5
H9JFU5
A0A2A4J427
A0A2H1VZ60
A0A194PCK6
A0A212ETK6
+ More
A0A0N1IPE4 K7J9K8 A0A069DVD0 A0A023F8T3 A0A088AP45 A0A195CAB7 F4X412 A0A151X6E4 V9IGE9 A0A195BGZ4 A0A195FPB6 A0A1Y1LJQ3 A0A154PNW3 U5EYE4 A0A067QJ19 E2B3Y8 A0A0M8ZVN2 E9JC37 A0A2A3EQB1 A0A1L8DVM6 A0A0A9Y262 A0A310SDV2 A0A1B6F339 A0A1B0CC27 A0A026WD67 E2ARX2 A0A158NJL0 A0A0P4VUW5 R4FNW4 Q178J5 A0A084VFJ0 A0A1B6IS61 D2A3J6 A0A023EUH7 A0A1J1J681 A0A182KG01 A0A182WYD0 A0A182U4T0 Q7PSJ1 A0A182KVP4 A0A182I5F8 A0A182YRK4 A0A182V256 A0A182PJD2 A0A182GMC6 A0A182NC68 A0A182IZY4 A0A2M4A4A8 A0A182RW26 B0WDS3 A0A2M4A4A5 A0A182QWA7 W5JGQ4 A0A1Q3FHJ0 A0A1Q3FHM5 A0A0K8TMF5 A0A0L7QSX7 A0A2M4BH03 A0A336MT09 A0A2M4BGY1 A0A2M4BGY6 J3JTJ5 A0A336MV84 A0A182F467 A0A2M3Z0M9 A0A182M497 A0A1W4XHC4 A0A1S3DAA0 A0A2S2QQE9 A0A0C9PLB3 A0A2S2P6A5 A0A2H8U1X4 J9K9M6 A0A2R7WSB6 A0A1Z5KVX9 E0VIM1 A0A162D7B5 A0A2R5LN40 A0A0P5XM93 A0A0P4Y2R0 E9G6J2 A0A0P5A1J5 A0A0P4XZY5 A0A0P5A2K5 A0A293M1B2 A0A0K8RIY4 A0A0P6E389 A0A0P5ZH84 A0A2L2Y3W6 A0A0P5MS51 A0A087UZW9 A0A023GD26 A0A1S3JUS1 A0A131XEV6
A0A0N1IPE4 K7J9K8 A0A069DVD0 A0A023F8T3 A0A088AP45 A0A195CAB7 F4X412 A0A151X6E4 V9IGE9 A0A195BGZ4 A0A195FPB6 A0A1Y1LJQ3 A0A154PNW3 U5EYE4 A0A067QJ19 E2B3Y8 A0A0M8ZVN2 E9JC37 A0A2A3EQB1 A0A1L8DVM6 A0A0A9Y262 A0A310SDV2 A0A1B6F339 A0A1B0CC27 A0A026WD67 E2ARX2 A0A158NJL0 A0A0P4VUW5 R4FNW4 Q178J5 A0A084VFJ0 A0A1B6IS61 D2A3J6 A0A023EUH7 A0A1J1J681 A0A182KG01 A0A182WYD0 A0A182U4T0 Q7PSJ1 A0A182KVP4 A0A182I5F8 A0A182YRK4 A0A182V256 A0A182PJD2 A0A182GMC6 A0A182NC68 A0A182IZY4 A0A2M4A4A8 A0A182RW26 B0WDS3 A0A2M4A4A5 A0A182QWA7 W5JGQ4 A0A1Q3FHJ0 A0A1Q3FHM5 A0A0K8TMF5 A0A0L7QSX7 A0A2M4BH03 A0A336MT09 A0A2M4BGY1 A0A2M4BGY6 J3JTJ5 A0A336MV84 A0A182F467 A0A2M3Z0M9 A0A182M497 A0A1W4XHC4 A0A1S3DAA0 A0A2S2QQE9 A0A0C9PLB3 A0A2S2P6A5 A0A2H8U1X4 J9K9M6 A0A2R7WSB6 A0A1Z5KVX9 E0VIM1 A0A162D7B5 A0A2R5LN40 A0A0P5XM93 A0A0P4Y2R0 E9G6J2 A0A0P5A1J5 A0A0P4XZY5 A0A0P5A2K5 A0A293M1B2 A0A0K8RIY4 A0A0P6E389 A0A0P5ZH84 A0A2L2Y3W6 A0A0P5MS51 A0A087UZW9 A0A023GD26 A0A1S3JUS1 A0A131XEV6
PDB
6FRK
E-value=0,
Score=1657
Ontologies
GO
Topology
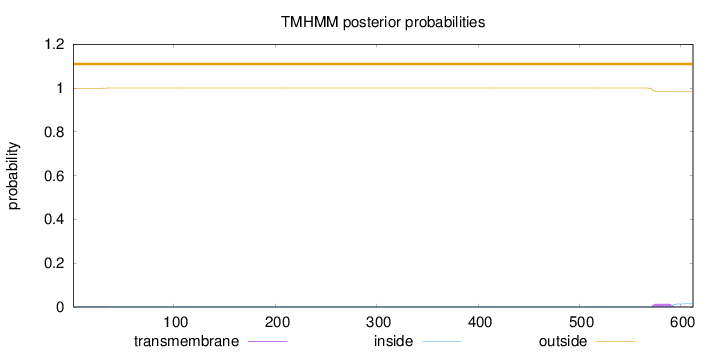
Length:
612
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.31243
Exp number, first 60 AAs:
0.0161
Total prob of N-in:
0.00093
outside
1 - 612
Population Genetic Test Statistics
Pi
189.363953
Theta
18.769357
Tajima's D
-1.22658
CLR
368.213093
CSRT
0.0996450177491125
Interpretation
Uncertain
