Pre Gene Modal
BGIBMGA008424
Annotation
PREDICTED:_protein_maelstrom_homolog_[Bombyx_mori]
Full name
Protein maelstrom homolog
Transcription factor
Location in the cell
Nuclear Reliability : 2.953
Sequence
CDS
ATGCCAAAAAAGAAGACCCCACAAAATCCATTTTTCTTCTTCATGATGGACTATAGGAAAGAACAAGCAGAAATAGGCATAAAATATGCCAACACCAAGGAATTAGCTGAAGCCGCTGGTCCTGTATGGCAGAACTTAAGGCCTACATTAAAAGCAAAATATGAAGAGAAAGCTAAGAATGAAAAAGAAAAGAACAAACAAACTGGAACTAAGTTCACGTCGACCGGCATACCTATAAAAGTAATAGAACAACAACAGAGAGAAATGAAGAATGCTGAAGATAATGAAAAAAAAGACATACAGAATATTGTGAAGCTCAAGGTGTTTGATCAAAGTATAAAAACTGAAGATTTTTATGTGATTGACGTGAACTCATACTGCAAAGCTAATGGTGATTATTTAATCGGAGAGTTCACCGTGACTCAATTCAGTCTGCAGGATGGTGTCAAAAATAGCTACCATGAAACTATTATTCCAAGTTGTGTACCGGTTGGATATATGTTTGATGTGAAACTGGGTGCTGAAGAGTTCGGCCTGGAGATGCCCGGCACCGATGACGCAGGACCCAACTACATACAAATACTGGCCAACATCATCGACTACTTGAAGCAGAAGGATCGGACTGTGCAAGTCTTGCCACCGATGTTCACGTTGCCGGAGAAGGTGGACGCAGTGCAGAACTTCATCTCACAGATGTGTAATTGTGCGACCGAAGACGACTCGCTGTTCCGTATATACAAACTAGACACGTTTTTCTTCACTCTGATCAACGCTATCAGCAGTCATCACGACGAGGGCTTCCCTAAGGAGTCGTTGGCGCTCACGCAGCTGACCAAGGACCTGTTCGACTACACACCGGGGATCGCGTGTGAGCGTCACGAGTCGCTGGACAGAAGCAACGTTTGCACAACGTCCCGCGTCAAACGTTGGGTGTTCACGATCCTGGACCGCTGCTGCCCGCTACTGGGGATTCCCCTGCAGCCGGGGAAGCACCTGCCCTTCGACTACGACATCAACGGCATCCTGATTTTTAAAGAAGAAAGGAAGACAAGAGCCGCTCCCTCTGTGCCTCGTACGGCCACCGCCTCGAGTAACTCATCCTTTATCAACGACTCTTTGAATGAATCCTTCCAAAGCTTGAACGTTACCGGCGAGACCGCCGCGTCGGGCTCGGCGCCGGGCAGGAGGGTGCACAAGCCGCTGCGCATGCCGCGTACCGACTATTCTCAGCGGATTCAGCAGGCTCCCGAACTGACGGAGGCTAACTTCCCGACGCTCACGGGGCACGGCCGAGGGCGTGGTCTGACCAGGAGCAACAATTGA
Protein
MPKKKTPQNPFFFFMMDYRKEQAEIGIKYANTKELAEAAGPVWQNLRPTLKAKYEEKAKNEKEKNKQTGTKFTSTGIPIKVIEQQQREMKNAEDNEKKDIQNIVKLKVFDQSIKTEDFYVIDVNSYCKANGDYLIGEFTVTQFSLQDGVKNSYHETIIPSCVPVGYMFDVKLGAEEFGLEMPGTDDAGPNYIQILANIIDYLKQKDRTVQVLPPMFTLPEKVDAVQNFISQMCNCATEDDSLFRIYKLDTFFFTLINAISSHHDEGFPKESLALTQLTKDLFDYTPGIACERHESLDRSNVCTTSRVKRWVFTILDRCCPLLGIPLQPGKHLPFDYDINGILIFKEERKTRAAPSVPRTATASSNSSFINDSLNESFQSLNVTGETAASGSAPGRRVHKPLRMPRTDYSQRIQQAPELTEANFPTLTGHGRGRGLTRSNN
Summary
Description
Plays a central role during gametogenesis by repressing transposable elements and preventing their mobilization, which is essential for the germline integrity. Probably acts via the piRNA metabolic process, which mediates the repression of transposable elements during meiosis by forming complexes composed of piRNAs and Piwi proteins and governs the repression of transposons (By similarity).
Similarity
Belongs to the maelstrom family.
Keywords
Complete proteome
Cytoplasm
Developmental protein
Differentiation
DNA-binding
Meiosis
Nucleus
Reference proteome
RNA-mediated gene silencing
Feature
chain Protein maelstrom homolog
Uniprot
A0A0H5AXR2
A0A212ETL5
S4PDX9
A0A3S2LSC5
A0A194PE20
A0A0N1IP66
+ More
A0A2H1WIG8 A0A2P8Z899 A0A2J7PG07 A0A067QTJ5 A0A2P8YWD1 A0A182PRJ3 A0A069DU44 A0A1B6J7Z9 A0A224XJV4 B0W2W6 A0A1B0EZW0 A0A1B0D1X9 B0W494 A0A2P8Z8A9 A0A1B6G106 A0A0V0G7H6 A0A1Q3F079 A0A1Q3F023 E0VM85 A0A1Q3F073 A0A1Q3EZN9 A0A182Q4Y0 A0A182I042 A0A182LH32 Q7PYM9 A0A1S4G6Y0 A0A1B6KNM8 A0A0P4VWA8 A0A182G7Y4 A0A182XBA0 A0A182KB73 A0A182V656 A0A182U671 A0A2M3ZJN9 A0A182G7Y5 A0A023EXL8 A0A1W4WEJ8 A0A1B0CVR1 A0A182FNR3 A0A2M4AWW2 A0A2M4AXC4 A0A2M4BLZ2 A0A2M4BLY9 A0A2M4BMU7 A0A1L8DAP5 A0A0A9Y2N6 A0A1L8DB70 V5GTW6 A0A0T6B2H4 W5JJY8 A0A1I8PJ21 A0A182H175 A0A182IIY3 A0A182NCG6 A0A182IKG6 U5EVE7 A0A084VJ51 T1H9A4 A0A1Q3EW40 Q16YA8 A0A170YYN4 A0A1I8PQG2 A0A182W4I5 A0A1B6CQG3 A0A0K8VZK8 A0A084VHB0 N6U553 A0A084VTZ5 A0A2J7QTC0 U4U4D0 A0A026WCS1 A0A1A9WKI0 A0A1Y1LH61 A0A0C9R9V5 A0A154PA32 T1PAS7 A0A182S6Z3 A0A2J7PV15 A0A034WUS8 A0A182YJT8 A0A1B0D8T9 A0A088ASY7 A0A310SH33 A0A1I8MZI2 A0A1B0B350 A0A182R9G6 A0A1A9ZXQ9
A0A2H1WIG8 A0A2P8Z899 A0A2J7PG07 A0A067QTJ5 A0A2P8YWD1 A0A182PRJ3 A0A069DU44 A0A1B6J7Z9 A0A224XJV4 B0W2W6 A0A1B0EZW0 A0A1B0D1X9 B0W494 A0A2P8Z8A9 A0A1B6G106 A0A0V0G7H6 A0A1Q3F079 A0A1Q3F023 E0VM85 A0A1Q3F073 A0A1Q3EZN9 A0A182Q4Y0 A0A182I042 A0A182LH32 Q7PYM9 A0A1S4G6Y0 A0A1B6KNM8 A0A0P4VWA8 A0A182G7Y4 A0A182XBA0 A0A182KB73 A0A182V656 A0A182U671 A0A2M3ZJN9 A0A182G7Y5 A0A023EXL8 A0A1W4WEJ8 A0A1B0CVR1 A0A182FNR3 A0A2M4AWW2 A0A2M4AXC4 A0A2M4BLZ2 A0A2M4BLY9 A0A2M4BMU7 A0A1L8DAP5 A0A0A9Y2N6 A0A1L8DB70 V5GTW6 A0A0T6B2H4 W5JJY8 A0A1I8PJ21 A0A182H175 A0A182IIY3 A0A182NCG6 A0A182IKG6 U5EVE7 A0A084VJ51 T1H9A4 A0A1Q3EW40 Q16YA8 A0A170YYN4 A0A1I8PQG2 A0A182W4I5 A0A1B6CQG3 A0A0K8VZK8 A0A084VHB0 N6U553 A0A084VTZ5 A0A2J7QTC0 U4U4D0 A0A026WCS1 A0A1A9WKI0 A0A1Y1LH61 A0A0C9R9V5 A0A154PA32 T1PAS7 A0A182S6Z3 A0A2J7PV15 A0A034WUS8 A0A182YJT8 A0A1B0D8T9 A0A088ASY7 A0A310SH33 A0A1I8MZI2 A0A1B0B350 A0A182R9G6 A0A1A9ZXQ9
Pubmed
EMBL
LC032360
BAR94546.1
AGBW02012556
OWR44833.1
GAIX01004767
JAA87793.1
+ More
RSAL01000295 RVE42830.1 KQ459606 KPI90934.1 KQ460556 KPJ13996.1 ODYU01008884 SOQ52858.1 PYGN01000150 PSN52729.1 NEVH01025635 PNF15261.1 KK853289 KDR08911.1 PYGN01000323 PSN48553.1 GBGD01001454 JAC87435.1 GECU01012418 JAS95288.1 GFTR01006318 JAW10108.1 DS231829 AJVK01008640 AJVK01022452 AJVK01022453 DS231836 EDS33179.1 PSN52718.1 GECZ01013641 JAS56128.1 GECL01002738 JAP03386.1 GFDL01014091 JAV20954.1 GFDL01014142 JAV20903.1 DS235297 EEB14491.1 GFDL01014093 JAV20952.1 GFDL01014278 JAV20767.1 AXCN02000016 APCN01002042 AAAB01008987 EAA01399.5 GEBQ01026924 GEBQ01014586 GEBQ01012899 JAT13053.1 JAT25391.1 JAT27078.1 GDKW01000199 JAI56396.1 JXUM01047242 JXUM01047243 JXUM01047244 JXUM01047245 JXUM01047246 JXUM01047247 KQ561512 KXJ78325.1 GGFM01007990 MBW28741.1 JXUM01047249 KXJ78326.1 GAPW01000379 JAC13219.1 AJWK01031027 AJWK01031028 GGFK01011953 MBW45274.1 GGFK01011927 MBW45248.1 GGFJ01004959 MBW54100.1 GGFJ01004958 MBW54099.1 GGFJ01004957 MBW54098.1 GFDF01010674 JAV03410.1 GBHO01026062 GBHO01026061 GBHO01026060 GBHO01019819 GBRD01016798 GBRD01016797 GDHC01019645 GDHC01011782 GDHC01009800 GDHC01007297 GDHC01004372 JAG17542.1 JAG17543.1 JAG17544.1 JAG23785.1 JAG49029.1 JAP98983.1 JAQ06847.1 JAQ08829.1 JAQ11332.1 JAQ14257.1 GFDF01010412 JAV03672.1 GALX01003384 JAB65082.1 LJIG01016088 KRT81651.1 ADMH02001019 ETN64416.1 JXUM01102761 KQ564750 KXJ71798.1 GANO01001894 JAB57977.1 ATLV01013431 KE524855 KFB37995.1 ACPB03005809 GFDL01015559 JAV19486.1 CH477519 GEMB01002807 JAS00391.1 GEDC01021529 JAS15769.1 GDHF01008045 JAI44269.1 ATLV01013150 KE524842 KFB37354.1 APGK01049250 KB741146 ENN73697.1 ATLV01016572 ATLV01016573 KE525094 KFB41439.1 NEVH01011196 PNF31828.1 KB631697 ERL85441.1 KK107263 EZA53862.1 GEZM01055727 JAV72964.1 GBYB01003596 JAG73363.1 KQ434857 KZC08796.1 KA645255 AFP59884.1 NEVH01021187 PNF20150.1 GAKP01000865 JAC58087.1 AJVK01027729 KQ761208 OAD57991.1 JXJN01007770
RSAL01000295 RVE42830.1 KQ459606 KPI90934.1 KQ460556 KPJ13996.1 ODYU01008884 SOQ52858.1 PYGN01000150 PSN52729.1 NEVH01025635 PNF15261.1 KK853289 KDR08911.1 PYGN01000323 PSN48553.1 GBGD01001454 JAC87435.1 GECU01012418 JAS95288.1 GFTR01006318 JAW10108.1 DS231829 AJVK01008640 AJVK01022452 AJVK01022453 DS231836 EDS33179.1 PSN52718.1 GECZ01013641 JAS56128.1 GECL01002738 JAP03386.1 GFDL01014091 JAV20954.1 GFDL01014142 JAV20903.1 DS235297 EEB14491.1 GFDL01014093 JAV20952.1 GFDL01014278 JAV20767.1 AXCN02000016 APCN01002042 AAAB01008987 EAA01399.5 GEBQ01026924 GEBQ01014586 GEBQ01012899 JAT13053.1 JAT25391.1 JAT27078.1 GDKW01000199 JAI56396.1 JXUM01047242 JXUM01047243 JXUM01047244 JXUM01047245 JXUM01047246 JXUM01047247 KQ561512 KXJ78325.1 GGFM01007990 MBW28741.1 JXUM01047249 KXJ78326.1 GAPW01000379 JAC13219.1 AJWK01031027 AJWK01031028 GGFK01011953 MBW45274.1 GGFK01011927 MBW45248.1 GGFJ01004959 MBW54100.1 GGFJ01004958 MBW54099.1 GGFJ01004957 MBW54098.1 GFDF01010674 JAV03410.1 GBHO01026062 GBHO01026061 GBHO01026060 GBHO01019819 GBRD01016798 GBRD01016797 GDHC01019645 GDHC01011782 GDHC01009800 GDHC01007297 GDHC01004372 JAG17542.1 JAG17543.1 JAG17544.1 JAG23785.1 JAG49029.1 JAP98983.1 JAQ06847.1 JAQ08829.1 JAQ11332.1 JAQ14257.1 GFDF01010412 JAV03672.1 GALX01003384 JAB65082.1 LJIG01016088 KRT81651.1 ADMH02001019 ETN64416.1 JXUM01102761 KQ564750 KXJ71798.1 GANO01001894 JAB57977.1 ATLV01013431 KE524855 KFB37995.1 ACPB03005809 GFDL01015559 JAV19486.1 CH477519 GEMB01002807 JAS00391.1 GEDC01021529 JAS15769.1 GDHF01008045 JAI44269.1 ATLV01013150 KE524842 KFB37354.1 APGK01049250 KB741146 ENN73697.1 ATLV01016572 ATLV01016573 KE525094 KFB41439.1 NEVH01011196 PNF31828.1 KB631697 ERL85441.1 KK107263 EZA53862.1 GEZM01055727 JAV72964.1 GBYB01003596 JAG73363.1 KQ434857 KZC08796.1 KA645255 AFP59884.1 NEVH01021187 PNF20150.1 GAKP01000865 JAC58087.1 AJVK01027729 KQ761208 OAD57991.1 JXJN01007770
Proteomes
UP000007151
UP000283053
UP000053268
UP000053240
UP000245037
UP000235965
+ More
UP000027135 UP000075885 UP000002320 UP000092462 UP000009046 UP000075886 UP000075840 UP000075882 UP000007062 UP000069940 UP000249989 UP000076407 UP000075881 UP000075903 UP000075902 UP000192223 UP000092461 UP000069272 UP000000673 UP000095300 UP000075880 UP000075884 UP000030765 UP000015103 UP000008820 UP000075920 UP000019118 UP000030742 UP000053097 UP000091820 UP000076502 UP000095301 UP000075901 UP000076408 UP000005203 UP000092460 UP000075900 UP000092445
UP000027135 UP000075885 UP000002320 UP000092462 UP000009046 UP000075886 UP000075840 UP000075882 UP000007062 UP000069940 UP000249989 UP000076407 UP000075881 UP000075903 UP000075902 UP000192223 UP000092461 UP000069272 UP000000673 UP000095300 UP000075880 UP000075884 UP000030765 UP000015103 UP000008820 UP000075920 UP000019118 UP000030742 UP000053097 UP000091820 UP000076502 UP000095301 UP000075901 UP000076408 UP000005203 UP000092460 UP000075900 UP000092445
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A0H5AXR2
A0A212ETL5
S4PDX9
A0A3S2LSC5
A0A194PE20
A0A0N1IP66
+ More
A0A2H1WIG8 A0A2P8Z899 A0A2J7PG07 A0A067QTJ5 A0A2P8YWD1 A0A182PRJ3 A0A069DU44 A0A1B6J7Z9 A0A224XJV4 B0W2W6 A0A1B0EZW0 A0A1B0D1X9 B0W494 A0A2P8Z8A9 A0A1B6G106 A0A0V0G7H6 A0A1Q3F079 A0A1Q3F023 E0VM85 A0A1Q3F073 A0A1Q3EZN9 A0A182Q4Y0 A0A182I042 A0A182LH32 Q7PYM9 A0A1S4G6Y0 A0A1B6KNM8 A0A0P4VWA8 A0A182G7Y4 A0A182XBA0 A0A182KB73 A0A182V656 A0A182U671 A0A2M3ZJN9 A0A182G7Y5 A0A023EXL8 A0A1W4WEJ8 A0A1B0CVR1 A0A182FNR3 A0A2M4AWW2 A0A2M4AXC4 A0A2M4BLZ2 A0A2M4BLY9 A0A2M4BMU7 A0A1L8DAP5 A0A0A9Y2N6 A0A1L8DB70 V5GTW6 A0A0T6B2H4 W5JJY8 A0A1I8PJ21 A0A182H175 A0A182IIY3 A0A182NCG6 A0A182IKG6 U5EVE7 A0A084VJ51 T1H9A4 A0A1Q3EW40 Q16YA8 A0A170YYN4 A0A1I8PQG2 A0A182W4I5 A0A1B6CQG3 A0A0K8VZK8 A0A084VHB0 N6U553 A0A084VTZ5 A0A2J7QTC0 U4U4D0 A0A026WCS1 A0A1A9WKI0 A0A1Y1LH61 A0A0C9R9V5 A0A154PA32 T1PAS7 A0A182S6Z3 A0A2J7PV15 A0A034WUS8 A0A182YJT8 A0A1B0D8T9 A0A088ASY7 A0A310SH33 A0A1I8MZI2 A0A1B0B350 A0A182R9G6 A0A1A9ZXQ9
A0A2H1WIG8 A0A2P8Z899 A0A2J7PG07 A0A067QTJ5 A0A2P8YWD1 A0A182PRJ3 A0A069DU44 A0A1B6J7Z9 A0A224XJV4 B0W2W6 A0A1B0EZW0 A0A1B0D1X9 B0W494 A0A2P8Z8A9 A0A1B6G106 A0A0V0G7H6 A0A1Q3F079 A0A1Q3F023 E0VM85 A0A1Q3F073 A0A1Q3EZN9 A0A182Q4Y0 A0A182I042 A0A182LH32 Q7PYM9 A0A1S4G6Y0 A0A1B6KNM8 A0A0P4VWA8 A0A182G7Y4 A0A182XBA0 A0A182KB73 A0A182V656 A0A182U671 A0A2M3ZJN9 A0A182G7Y5 A0A023EXL8 A0A1W4WEJ8 A0A1B0CVR1 A0A182FNR3 A0A2M4AWW2 A0A2M4AXC4 A0A2M4BLZ2 A0A2M4BLY9 A0A2M4BMU7 A0A1L8DAP5 A0A0A9Y2N6 A0A1L8DB70 V5GTW6 A0A0T6B2H4 W5JJY8 A0A1I8PJ21 A0A182H175 A0A182IIY3 A0A182NCG6 A0A182IKG6 U5EVE7 A0A084VJ51 T1H9A4 A0A1Q3EW40 Q16YA8 A0A170YYN4 A0A1I8PQG2 A0A182W4I5 A0A1B6CQG3 A0A0K8VZK8 A0A084VHB0 N6U553 A0A084VTZ5 A0A2J7QTC0 U4U4D0 A0A026WCS1 A0A1A9WKI0 A0A1Y1LH61 A0A0C9R9V5 A0A154PA32 T1PAS7 A0A182S6Z3 A0A2J7PV15 A0A034WUS8 A0A182YJT8 A0A1B0D8T9 A0A088ASY7 A0A310SH33 A0A1I8MZI2 A0A1B0B350 A0A182R9G6 A0A1A9ZXQ9
PDB
5AF0
E-value=5.63486e-154,
Score=1396
Ontologies
GO
PANTHER
Topology
Subcellular location
Cytoplasm
Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis. With evidence from 1 publications.
Nucleus Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis. With evidence from 1 publications.
Nucleus Component of the meiotic nuage, also named P granule, a germ-cell-specific organelle required to repress transposon activity during meiosis. With evidence from 1 publications.
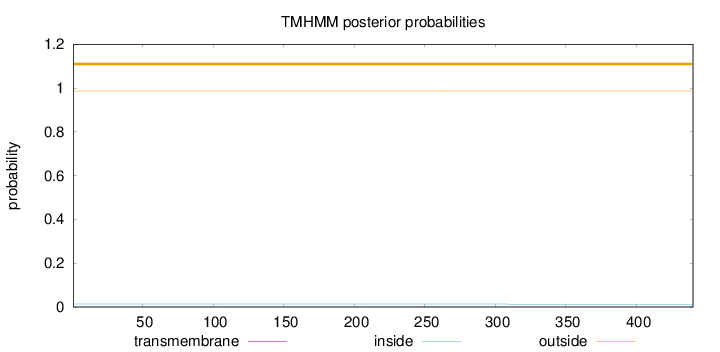
Length:
440
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00450999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01335
outside
1 - 440
Population Genetic Test Statistics
Pi
189.368412
Theta
143.037444
Tajima's D
0.590922
CLR
0.131723
CSRT
0.537973101344933
Interpretation
Uncertain
