Gene
KWMTBOMO10776
Pre Gene Modal
BGIBMGA008394
Annotation
PREDICTED:_alpha-(1?6)-fucosyltransferase_[Papilio_xuthus]
Full name
Alpha-(1,6)-fucosyltransferase
Location in the cell
Cytoplasmic Reliability : 1.192 Nuclear Reliability : 1.261
Sequence
CDS
ATGAAGAATTATTCAAATGGTCACGTAACGATACATGATTATTTTATGATGTACTTTGCGAAATGGAAGCGAGTCGCCGTCGTATTGCTCGCCGTTTGGATTGTCGTCACCTATTTAGTCATATCGCCTTTAAGATGCAGCGGCAATTCAGATGAGATCCCCGACGTCCAGGAGAGGTTGAAGCGGATGTCTTCGGAGCTGGAAGTCCTGTGGCAGAGAAACAGTAAGCTCATAGCGCAGATAAAGAAGTCTTCAGGGCCCAATGGCAATCTTAAAGATTTGGATCCGGCGTTTTGGAACCTTGAAGCGGGACTTGGTCCGTCAGAGGAATACGAGAATCTCAGGAGGAGGATCCACTCGAATACAAAGGAGTTCTGGTACTACGCGAACCATGAGCTCGGGAAGTTGATGCACGAGAGTGACAAGGAAGCGAAAATACGAGCGATTTTGGAACAACTGTCTGATAGGAAAAGCACATTACTGTCTGATCAAGAAAAACTATCGGAAATGGATGGTTACCACGAGTGGAGGCAACTAGAGGCGGCCAACGTCAGCGACCTTGTCCAGAGGAGGTTGCACTATTTACAGAACCCGCCGGATTGTAGGGAGGCAAGGAAACTTATATGCAACTTGAATAAGGGCTGCGGCTTCGGCTGTCAACTCCACCACATAGTGTACTGCCTTATATTCGCTTACGCCACCGAGAGGACTCTAATCCTGAACTCGAAGGGGTGGCGGTACAACACTAAAGGATGGGAGTACGTCTTCCATCCGATATCGGAGAGCTGTTTGTCTTCGTACGACGATAAAGTCGTGCAGTGGCCAGTTTCATACGACGCGAAAGTGGTTTCGCTACCGTTTATAGATTCCATATCGCAGAAGCCCAAGTTCTTGCCGCTGGCTGTGCCTAAAGACTTGGCTCATAGGATAGTCCGCTTCAACGGCGATCCGTCGTCGTGGTGGGTCGGGCAAATGCTGAAGTTCGTCCTGAAGCCGCGCCTGCCGATGCAGAAGGCGATCAACGACACAATAGCCAAAATGAATTTCAAATCTCCCATAGTCGGGGTTCACATACGCCGCACCGATAAAGTTGGAACCGAAGCCGCCTTCCACCACATACACGAGTACATGGCGCACGTGAAGGACTATTACGACCAGCTAGAGCTGACCCGGCCCGTCGACGTCCGGAGAGTGTATCTAGCCACAGACGACGCTAATGTGCTAGACGACGCCCGACAAAAGTATCCGGAGTACACGTTCTTAGGGGATCCGTCGATAGCGAAGACGGCGGCCACTCACCGTAGATACACGCCGCTCTCGCTTACTGGCCTACTGGTGGATCTACACATGCTGGCCATGTGCGATTACTTAGTGTGTACATTTAGCAGTCAAGTGGGTCGAGTGGCGTACGAAATGATGCAGTCGAACAGAGTGGACGCGTCAAACAGCTTCTTCTCGTTAGATGACATTTACTACTTCGGTGGACAAAACGCGCACGACAGGGTGGCCATCATGCAGAACCACGGCGGGAAAAACGAGGATATATCTTTTGAGGTGGGCGATAAAATAGGAGTGGCCGGCAACCATTGGAACGGATACGGACGCGGAACTAACAAGAGAACGAATATGAACGGTCTGATCCCTTGGTACAAGACTGCCGATCATCTGGTGTTGTACCCGTTTCCGGAATACAAACAGGTGCCTATTTATTCAGAAACGAGCCAAAAGAACGTTTAA
Protein
MKNYSNGHVTIHDYFMMYFAKWKRVAVVLLAVWIVVTYLVISPLRCSGNSDEIPDVQERLKRMSSELEVLWQRNSKLIAQIKKSSGPNGNLKDLDPAFWNLEAGLGPSEEYENLRRRIHSNTKEFWYYANHELGKLMHESDKEAKIRAILEQLSDRKSTLLSDQEKLSEMDGYHEWRQLEAANVSDLVQRRLHYLQNPPDCREARKLICNLNKGCGFGCQLHHIVYCLIFAYATERTLILNSKGWRYNTKGWEYVFHPISESCLSSYDDKVVQWPVSYDAKVVSLPFIDSISQKPKFLPLAVPKDLAHRIVRFNGDPSSWWVGQMLKFVLKPRLPMQKAINDTIAKMNFKSPIVGVHIRRTDKVGTEAAFHHIHEYMAHVKDYYDQLELTRPVDVRRVYLATDDANVLDDARQKYPEYTFLGDPSIAKTAATHRRYTPLSLTGLLVDLHMLAMCDYLVCTFSSQVGRVAYEMMQSNRVDASNSFFSLDDIYYFGGQNAHDRVAIMQNHGGKNEDISFEVGDKIGVAGNHWNGYGRGTNKRTNMNGLIPWYKTADHLVLYPFPEYKQVPIYSETSQKNV
Summary
Description
Catalyzes the addition of fucose in alpha 1-6 linkage to the first GlcNAc residue, next to the peptide chains in N-glycans.
Catalytic Activity
GDP-beta-L-fucose + N(4)-{beta-D-GlcNAc-(1->2)-alpha-D-Man-(1->3)-[beta-D-GlcNAc-(1->2)-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAc-(1->4)-beta-D-GlcNAc}-L-asparaginyl-[protein] = GDP + H(+) + N(4)-{beta-D-GlcNAc-(1->2)-alpha-D-Man-(1->3)-[beta-D-GlcNAc-(1->2)-alpha-D-Man-(1->6)]-beta-D-Man-(1->4)-beta-D-GlcNAcl-(1->4)-[alpha-L-Fuc-(1->6)]-beta-D-GlcNAc}-L-asparaginyl-[protein]
Similarity
Belongs to the glycosyltransferase 23 family.
Feature
chain Alpha-(1,6)-fucosyltransferase
Uniprot
A0A060PLD2
H9JFU7
A0A2A4JPZ2
A0A194PIL8
R4L884
A0A0N1I7K9
+ More
A0A212ETJ9 A0A0A1TKV6 A0A1Y1L2Q9 A0A139WIJ8 D2A2U6 A0A182JQS5 A0A1B6CPN6 A0A2P8YME2 A0A182MUF6 A0A182VCR0 A0A182XBM1 A0A182TYT5 A0A182I0F5 Q7PYD8 A0A2J7QLV9 A0A0P4Z061 A0A0P6HXG2 A0A164L1R6 E9HF16 A0A0A1TKW5 A0A182YJU6 B4MAI6 B4L679 A0A182SAJ5 A0A182Q8X1 A0A1Q3FF33 A0A067RGX4 A0A182INY9 A0A182PC20 A0A0M4EML8 A0A182N5X1 A0A1L8E215 A0A023EV61 U5EZ81 B0WME2 A0A182G1U2 A0A336LNK4 A0A1L8E2C2 N6T2L9 B4MT17 A0A1L8E233 A0A2M4A0D1 A0A034VSZ3 A0A2S2QB72 A0A084VRW7 W5J3H7 A0A0P6DDA9 A0A0A1X468 A0A2M3YYP9 A0A2M3ZDZ8 A0A182FQK7 A0A2M4BHX1 A0A2S2QAD6 Q16ZK5 A0A1W4XMA5 A0A0K8UIV4 A0A182VQX6 W8BPV9 A0A1J1IY37 A0A2M4BHX2 A0A2S2P711 A0A0P5W4L6 B4JM16 A0A087SYF1 A0A1I8M9H5 A0A1A9YMH3 A0A1B0BDK8 A0A1B0FAZ8 X1XL71 A0A0A1TTF2 A0A2H8TE61 A0A0P5TI54 A0A1B0A828 A0A0L0BVR5 A0A1A9WVK8 A0A182KM43 A0A293MSG4 A0A1B0GKV9 A0A1I8NTE8 A0A1A9UW33 A0A0N0BIS4 A0A1B6KQG1 W4WWN8 A0A2A3EDG2 A0A195CJ35 A0A195F151 A0A1B6JXJ5 A0A195B2I4 A0A182R1I4 A0A195E6L9 F4X6S2 A0A088AHL8 A0A1W4WBG7 A0A0L7RIK4
A0A212ETJ9 A0A0A1TKV6 A0A1Y1L2Q9 A0A139WIJ8 D2A2U6 A0A182JQS5 A0A1B6CPN6 A0A2P8YME2 A0A182MUF6 A0A182VCR0 A0A182XBM1 A0A182TYT5 A0A182I0F5 Q7PYD8 A0A2J7QLV9 A0A0P4Z061 A0A0P6HXG2 A0A164L1R6 E9HF16 A0A0A1TKW5 A0A182YJU6 B4MAI6 B4L679 A0A182SAJ5 A0A182Q8X1 A0A1Q3FF33 A0A067RGX4 A0A182INY9 A0A182PC20 A0A0M4EML8 A0A182N5X1 A0A1L8E215 A0A023EV61 U5EZ81 B0WME2 A0A182G1U2 A0A336LNK4 A0A1L8E2C2 N6T2L9 B4MT17 A0A1L8E233 A0A2M4A0D1 A0A034VSZ3 A0A2S2QB72 A0A084VRW7 W5J3H7 A0A0P6DDA9 A0A0A1X468 A0A2M3YYP9 A0A2M3ZDZ8 A0A182FQK7 A0A2M4BHX1 A0A2S2QAD6 Q16ZK5 A0A1W4XMA5 A0A0K8UIV4 A0A182VQX6 W8BPV9 A0A1J1IY37 A0A2M4BHX2 A0A2S2P711 A0A0P5W4L6 B4JM16 A0A087SYF1 A0A1I8M9H5 A0A1A9YMH3 A0A1B0BDK8 A0A1B0FAZ8 X1XL71 A0A0A1TTF2 A0A2H8TE61 A0A0P5TI54 A0A1B0A828 A0A0L0BVR5 A0A1A9WVK8 A0A182KM43 A0A293MSG4 A0A1B0GKV9 A0A1I8NTE8 A0A1A9UW33 A0A0N0BIS4 A0A1B6KQG1 W4WWN8 A0A2A3EDG2 A0A195CJ35 A0A195F151 A0A1B6JXJ5 A0A195B2I4 A0A182R1I4 A0A195E6L9 F4X6S2 A0A088AHL8 A0A1W4WBG7 A0A0L7RIK4
EC Number
2.4.1.68
Pubmed
EMBL
AB848715
BAO96240.1
BABH01001589
BABH01001590
BABH01001591
NWSH01000842
+ More
PCG73889.1 KQ459606 KPI90935.1 KC538901 KC538902 AGL08712.1 KQ460556 KPJ13995.1 AGBW02012556 OWR44832.1 BK008802 DAA64658.1 GEZM01066303 JAV67979.1 KQ971339 KYB27749.1 EFA02784.1 GEDC01021851 JAS15447.1 PYGN01000494 PSN45403.1 AXCM01000371 APCN01002074 AAAB01008987 EAA01061.5 NEVH01013241 PNF29584.1 GDIP01219931 JAJ03471.1 GDIQ01014149 JAN80588.1 LRGB01003216 KZS03721.1 GL732633 EFX69666.1 BK008814 DAA64668.1 CH940655 EDW66245.1 CH933812 EDW05875.1 AXCN02000190 GFDL01008890 JAV26155.1 KK852478 KDR23027.1 CP012528 ALC49021.1 GFDF01001314 JAV12770.1 GAPW01000717 JAC12881.1 GANO01001268 JAB58603.1 DS231997 EDS30999.1 JXUM01006333 JXUM01006334 KQ560213 KXJ83815.1 UFQT01000095 SSX19724.1 GFDF01001315 JAV12769.1 APGK01046982 APGK01046983 KB741073 ENN74379.1 CH963851 EDW75256.1 GFDF01001313 JAV12771.1 GGFK01000955 MBW34276.1 GAKP01014289 JAC44663.1 GGMS01005784 MBY74987.1 ATLV01015783 KE525036 KFB40711.1 ADMH02002134 ETN58391.1 GDIQ01088149 JAN06588.1 GBXI01008829 JAD05463.1 GGFM01000643 MBW21394.1 GGFM01005957 MBW26708.1 GGFJ01003518 MBW52659.1 GGMS01005495 MBY74698.1 CH477489 EAT40069.1 GDHF01025677 GDHF01025507 GDHF01020054 JAI26637.1 JAI26807.1 JAI32260.1 GAMC01015052 GAMC01015051 JAB91504.1 CVRI01000063 CRL04476.1 GGFJ01003519 MBW52660.1 GGMR01012595 MBY25214.1 GDIP01104950 JAL98764.1 CH916371 EDV91777.1 KK112528 KFM57890.1 JXJN01012514 CCAG010001425 ABLF02016140 ABLF02016142 BK008810 DAA64664.1 GFXV01000581 MBW12386.1 GDIP01125955 JAL77759.1 JRES01001264 KNC24088.1 GFWV01019004 MAA43732.1 AJWK01033779 KQ435727 KOX77864.1 GEBQ01026302 GEBQ01001618 JAT13675.1 JAT38359.1 BK008808 DAA64662.1 KZ288282 PBC29524.1 KQ977754 KYN00079.1 KQ981864 KYN34295.1 GECU01003780 JAT03927.1 KQ976662 KYM78502.1 KQ979568 KYN20840.1 GL888818 EGI57854.1 KQ414584 KOC70561.1
PCG73889.1 KQ459606 KPI90935.1 KC538901 KC538902 AGL08712.1 KQ460556 KPJ13995.1 AGBW02012556 OWR44832.1 BK008802 DAA64658.1 GEZM01066303 JAV67979.1 KQ971339 KYB27749.1 EFA02784.1 GEDC01021851 JAS15447.1 PYGN01000494 PSN45403.1 AXCM01000371 APCN01002074 AAAB01008987 EAA01061.5 NEVH01013241 PNF29584.1 GDIP01219931 JAJ03471.1 GDIQ01014149 JAN80588.1 LRGB01003216 KZS03721.1 GL732633 EFX69666.1 BK008814 DAA64668.1 CH940655 EDW66245.1 CH933812 EDW05875.1 AXCN02000190 GFDL01008890 JAV26155.1 KK852478 KDR23027.1 CP012528 ALC49021.1 GFDF01001314 JAV12770.1 GAPW01000717 JAC12881.1 GANO01001268 JAB58603.1 DS231997 EDS30999.1 JXUM01006333 JXUM01006334 KQ560213 KXJ83815.1 UFQT01000095 SSX19724.1 GFDF01001315 JAV12769.1 APGK01046982 APGK01046983 KB741073 ENN74379.1 CH963851 EDW75256.1 GFDF01001313 JAV12771.1 GGFK01000955 MBW34276.1 GAKP01014289 JAC44663.1 GGMS01005784 MBY74987.1 ATLV01015783 KE525036 KFB40711.1 ADMH02002134 ETN58391.1 GDIQ01088149 JAN06588.1 GBXI01008829 JAD05463.1 GGFM01000643 MBW21394.1 GGFM01005957 MBW26708.1 GGFJ01003518 MBW52659.1 GGMS01005495 MBY74698.1 CH477489 EAT40069.1 GDHF01025677 GDHF01025507 GDHF01020054 JAI26637.1 JAI26807.1 JAI32260.1 GAMC01015052 GAMC01015051 JAB91504.1 CVRI01000063 CRL04476.1 GGFJ01003519 MBW52660.1 GGMR01012595 MBY25214.1 GDIP01104950 JAL98764.1 CH916371 EDV91777.1 KK112528 KFM57890.1 JXJN01012514 CCAG010001425 ABLF02016140 ABLF02016142 BK008810 DAA64664.1 GFXV01000581 MBW12386.1 GDIP01125955 JAL77759.1 JRES01001264 KNC24088.1 GFWV01019004 MAA43732.1 AJWK01033779 KQ435727 KOX77864.1 GEBQ01026302 GEBQ01001618 JAT13675.1 JAT38359.1 BK008808 DAA64662.1 KZ288282 PBC29524.1 KQ977754 KYN00079.1 KQ981864 KYN34295.1 GECU01003780 JAT03927.1 KQ976662 KYM78502.1 KQ979568 KYN20840.1 GL888818 EGI57854.1 KQ414584 KOC70561.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000007266
+ More
UP000075881 UP000245037 UP000075883 UP000075903 UP000076407 UP000075902 UP000075840 UP000007062 UP000235965 UP000076858 UP000000305 UP000076408 UP000008792 UP000009192 UP000075901 UP000075886 UP000027135 UP000075880 UP000075885 UP000092553 UP000075884 UP000002320 UP000069940 UP000249989 UP000019118 UP000007798 UP000030765 UP000000673 UP000069272 UP000008820 UP000192223 UP000075920 UP000183832 UP000001070 UP000054359 UP000095301 UP000092443 UP000092460 UP000092444 UP000007819 UP000092445 UP000037069 UP000091820 UP000075882 UP000092461 UP000095300 UP000078200 UP000053105 UP000005205 UP000242457 UP000078542 UP000078541 UP000078540 UP000075900 UP000078492 UP000007755 UP000005203 UP000192221 UP000053825
UP000075881 UP000245037 UP000075883 UP000075903 UP000076407 UP000075902 UP000075840 UP000007062 UP000235965 UP000076858 UP000000305 UP000076408 UP000008792 UP000009192 UP000075901 UP000075886 UP000027135 UP000075880 UP000075885 UP000092553 UP000075884 UP000002320 UP000069940 UP000249989 UP000019118 UP000007798 UP000030765 UP000000673 UP000069272 UP000008820 UP000192223 UP000075920 UP000183832 UP000001070 UP000054359 UP000095301 UP000092443 UP000092460 UP000092444 UP000007819 UP000092445 UP000037069 UP000091820 UP000075882 UP000092461 UP000095300 UP000078200 UP000053105 UP000005205 UP000242457 UP000078542 UP000078541 UP000078540 UP000075900 UP000078492 UP000007755 UP000005203 UP000192221 UP000053825
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A060PLD2
H9JFU7
A0A2A4JPZ2
A0A194PIL8
R4L884
A0A0N1I7K9
+ More
A0A212ETJ9 A0A0A1TKV6 A0A1Y1L2Q9 A0A139WIJ8 D2A2U6 A0A182JQS5 A0A1B6CPN6 A0A2P8YME2 A0A182MUF6 A0A182VCR0 A0A182XBM1 A0A182TYT5 A0A182I0F5 Q7PYD8 A0A2J7QLV9 A0A0P4Z061 A0A0P6HXG2 A0A164L1R6 E9HF16 A0A0A1TKW5 A0A182YJU6 B4MAI6 B4L679 A0A182SAJ5 A0A182Q8X1 A0A1Q3FF33 A0A067RGX4 A0A182INY9 A0A182PC20 A0A0M4EML8 A0A182N5X1 A0A1L8E215 A0A023EV61 U5EZ81 B0WME2 A0A182G1U2 A0A336LNK4 A0A1L8E2C2 N6T2L9 B4MT17 A0A1L8E233 A0A2M4A0D1 A0A034VSZ3 A0A2S2QB72 A0A084VRW7 W5J3H7 A0A0P6DDA9 A0A0A1X468 A0A2M3YYP9 A0A2M3ZDZ8 A0A182FQK7 A0A2M4BHX1 A0A2S2QAD6 Q16ZK5 A0A1W4XMA5 A0A0K8UIV4 A0A182VQX6 W8BPV9 A0A1J1IY37 A0A2M4BHX2 A0A2S2P711 A0A0P5W4L6 B4JM16 A0A087SYF1 A0A1I8M9H5 A0A1A9YMH3 A0A1B0BDK8 A0A1B0FAZ8 X1XL71 A0A0A1TTF2 A0A2H8TE61 A0A0P5TI54 A0A1B0A828 A0A0L0BVR5 A0A1A9WVK8 A0A182KM43 A0A293MSG4 A0A1B0GKV9 A0A1I8NTE8 A0A1A9UW33 A0A0N0BIS4 A0A1B6KQG1 W4WWN8 A0A2A3EDG2 A0A195CJ35 A0A195F151 A0A1B6JXJ5 A0A195B2I4 A0A182R1I4 A0A195E6L9 F4X6S2 A0A088AHL8 A0A1W4WBG7 A0A0L7RIK4
A0A212ETJ9 A0A0A1TKV6 A0A1Y1L2Q9 A0A139WIJ8 D2A2U6 A0A182JQS5 A0A1B6CPN6 A0A2P8YME2 A0A182MUF6 A0A182VCR0 A0A182XBM1 A0A182TYT5 A0A182I0F5 Q7PYD8 A0A2J7QLV9 A0A0P4Z061 A0A0P6HXG2 A0A164L1R6 E9HF16 A0A0A1TKW5 A0A182YJU6 B4MAI6 B4L679 A0A182SAJ5 A0A182Q8X1 A0A1Q3FF33 A0A067RGX4 A0A182INY9 A0A182PC20 A0A0M4EML8 A0A182N5X1 A0A1L8E215 A0A023EV61 U5EZ81 B0WME2 A0A182G1U2 A0A336LNK4 A0A1L8E2C2 N6T2L9 B4MT17 A0A1L8E233 A0A2M4A0D1 A0A034VSZ3 A0A2S2QB72 A0A084VRW7 W5J3H7 A0A0P6DDA9 A0A0A1X468 A0A2M3YYP9 A0A2M3ZDZ8 A0A182FQK7 A0A2M4BHX1 A0A2S2QAD6 Q16ZK5 A0A1W4XMA5 A0A0K8UIV4 A0A182VQX6 W8BPV9 A0A1J1IY37 A0A2M4BHX2 A0A2S2P711 A0A0P5W4L6 B4JM16 A0A087SYF1 A0A1I8M9H5 A0A1A9YMH3 A0A1B0BDK8 A0A1B0FAZ8 X1XL71 A0A0A1TTF2 A0A2H8TE61 A0A0P5TI54 A0A1B0A828 A0A0L0BVR5 A0A1A9WVK8 A0A182KM43 A0A293MSG4 A0A1B0GKV9 A0A1I8NTE8 A0A1A9UW33 A0A0N0BIS4 A0A1B6KQG1 W4WWN8 A0A2A3EDG2 A0A195CJ35 A0A195F151 A0A1B6JXJ5 A0A195B2I4 A0A182R1I4 A0A195E6L9 F4X6S2 A0A088AHL8 A0A1W4WBG7 A0A0L7RIK4
PDB
2DE0
E-value=1.0373e-138,
Score=1266
Ontologies
PATHWAY
GO
Topology
Subcellular location
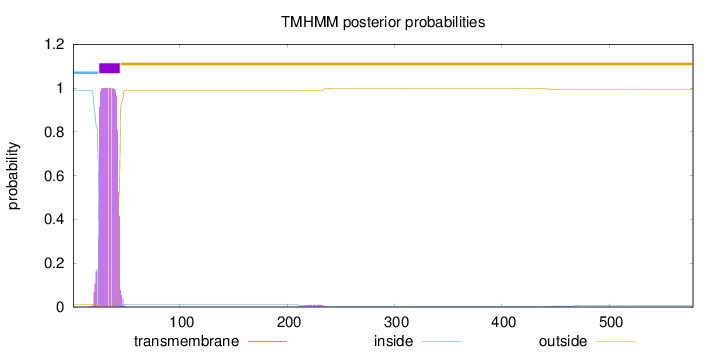
Length:
578
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.9231
Exp number, first 60 AAs:
19.60344
Total prob of N-in:
0.98941
POSSIBLE N-term signal
sequence
inside
1 - 24
TMhelix
25 - 44
outside
45 - 578
Population Genetic Test Statistics
Pi
243.012399
Theta
164.206865
Tajima's D
1.528415
CLR
0.375151
CSRT
0.793710314484276
Interpretation
Uncertain
