Gene
KWMTBOMO10773
Pre Gene Modal
BGIBMGA008395
Annotation
PREDICTED:_delta-like_protein_1_[Bombyx_mori]
Full name
Delta-like protein
+ More
Delta-like protein B
Delta-like protein C
Delta-like protein B
Delta-like protein C
Location in the cell
Extracellular Reliability : 2.136
Sequence
CDS
ATGTTGTCACTACCAAATTTAGAACCAATTTGTGCAGCGAACTGTCACAGCGAGAGGGGCTTCTGTCGGCGTCCCGGGGAATGCCGCTGCAAGGTCGGCTGGACCGGGCCCACGTGCGCGCAGTGCCACAGGTACCCTGGCTGCAAGCACGGAACTTGCAGGAAGCCTTGGGAGTGTATCTGCAAAAAGGGCTGGGGAGGAATGCTGTGTGACGAAGGTGAGTTTTAA
Protein
MLSLPNLEPICAANCHSERGFCRRPGECRCKVGWTGPTCAQCHRYPGCKHGTCRKPWECICKKGWGGMLCDEGEF
Summary
Description
Putative Notch ligand involved in the mediation of Notch signaling.
Acts as a ligand for Notch receptors and is involved in primary neurogenesis. Can activate Notch receptors, thereby playing a key role in lateral inhibition, a process that prevents the immediate neighbors of each nascent neural cell from simultaneously embarking on neural differentiation.
Acts as a ligand for Notch receptors and is involved in somitogenesis. Can activate Notch receptors. Required in somite segmentation to keep the oscillations of neighboring presomitic mesoderm cells synchronized.
Acts as a ligand for Notch receptors and is involved in primary neurogenesis. Can activate Notch receptors, thereby playing a key role in lateral inhibition, a process that prevents the immediate neighbors of each nascent neural cell from simultaneously embarking on neural differentiation.
Acts as a ligand for Notch receptors and is involved in somitogenesis. Can activate Notch receptors. Required in somite segmentation to keep the oscillations of neighboring presomitic mesoderm cells synchronized.
Keywords
Calcium
Complete proteome
Developmental protein
Differentiation
Disulfide bond
EGF-like domain
Glycoprotein
Membrane
Neurogenesis
Notch signaling pathway
Reference proteome
Repeat
Signal
Transmembrane
Transmembrane helix
Ubl conjugation
Feature
chain Delta-like protein
Uniprot
H9JFU8
A0A3S2NLG9
A0A2H1VJ88
A0A2A4JR51
A0A194PCC7
I4DJ83
+ More
A0A0N0PCG8 A0A212ETQ4 A0A2J7QLV8 A0A1I8PB87 A0A182NC65 W5JJZ3 A0A1B0DG68 A0A1I8MRK7 Q17IN2 A0A336LHW1 A0A1S4F1B8 A0A182WFV9 A0A0K8VB06 A0A0A1WMS8 A0A182RYW0 A0A034WVX6 A0A182WUN4 A0A182U5Y4 A0A182KCR2 A0A182KKC3 U4U1J1 A0A182UQG4 A0A182HVU1 A0A182PJ98 A0A182GGA6 A0A182FKD6 A0A182MDB1 D2A0G9 Q7QGY2 A0A067RJ30 A0A084WQA9 A0A182IWL1 A0A182YKN3 A0A182SV86 A0A2S2P900 A0A0K8W2B8 A0A161MR17 A0A1B6HDI0 A0A1S4N2N7 B0X8Q5 E0W3E6 A0A1B0A1B0 A0A1B6EQ11 A0A1B6LUF6 A0A1B0G9S4 W8B1I0 W8AXK5 A0A1A9VWM8 J9JWS8 A0A2S2Q6W0 J3JXN7 A0A2H8TZD5 U4UGD5 A0A3B1ISZ0 A0A3B1K7P8 W5KPC7 A0A1A9WWW1 A0A0L0C381 A0A1Y1KKZ3 A0A3B0K5Y3 B4H4T9 B4MSI0 A0A3P8YLV6 T1IPW5 Q29I00 A0A2D0QCF8 A0A3B4DQL0 W5KGW2 A0A3P8YLP6 A0A3B4VBM2 Q1LYD2 O57409 A0A3B4WWN9 A0A3B4VBN9 A0A3B4X6T1 A0A3N0Y074 A0A060YM88 B4PYV7 A0A1S3RLH3 A0A3Q3DVC3 A0A1S3QDA7 Q9VZ44 B4JXB2 G3Q9T1 B3NVC3 B4MAN3 L8HSS5 A0A1W4V2L9 A0A2R8QUE3 A4JYS0 Q9IAT6 A0A3Q1K3S9 A0A3Q2XGU1 A0A3B4BGD1
A0A0N0PCG8 A0A212ETQ4 A0A2J7QLV8 A0A1I8PB87 A0A182NC65 W5JJZ3 A0A1B0DG68 A0A1I8MRK7 Q17IN2 A0A336LHW1 A0A1S4F1B8 A0A182WFV9 A0A0K8VB06 A0A0A1WMS8 A0A182RYW0 A0A034WVX6 A0A182WUN4 A0A182U5Y4 A0A182KCR2 A0A182KKC3 U4U1J1 A0A182UQG4 A0A182HVU1 A0A182PJ98 A0A182GGA6 A0A182FKD6 A0A182MDB1 D2A0G9 Q7QGY2 A0A067RJ30 A0A084WQA9 A0A182IWL1 A0A182YKN3 A0A182SV86 A0A2S2P900 A0A0K8W2B8 A0A161MR17 A0A1B6HDI0 A0A1S4N2N7 B0X8Q5 E0W3E6 A0A1B0A1B0 A0A1B6EQ11 A0A1B6LUF6 A0A1B0G9S4 W8B1I0 W8AXK5 A0A1A9VWM8 J9JWS8 A0A2S2Q6W0 J3JXN7 A0A2H8TZD5 U4UGD5 A0A3B1ISZ0 A0A3B1K7P8 W5KPC7 A0A1A9WWW1 A0A0L0C381 A0A1Y1KKZ3 A0A3B0K5Y3 B4H4T9 B4MSI0 A0A3P8YLV6 T1IPW5 Q29I00 A0A2D0QCF8 A0A3B4DQL0 W5KGW2 A0A3P8YLP6 A0A3B4VBM2 Q1LYD2 O57409 A0A3B4WWN9 A0A3B4VBN9 A0A3B4X6T1 A0A3N0Y074 A0A060YM88 B4PYV7 A0A1S3RLH3 A0A3Q3DVC3 A0A1S3QDA7 Q9VZ44 B4JXB2 G3Q9T1 B3NVC3 B4MAN3 L8HSS5 A0A1W4V2L9 A0A2R8QUE3 A4JYS0 Q9IAT6 A0A3Q1K3S9 A0A3Q2XGU1 A0A3B4BGD1
Pubmed
19121390
26354079
22651552
22118469
20920257
23761445
+ More
25315136 17510324 25830018 25348373 20966253 23537049 26483478 18362917 19820115 12364791 14747013 17210077 24845553 24438588 25244985 20566863 24495485 22516182 25329095 26108605 28004739 17994087 25069045 15632085 9425132 12530964 24755649 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22751099 23594743 10585570 11100729 25463417
25315136 17510324 25830018 25348373 20966253 23537049 26483478 18362917 19820115 12364791 14747013 17210077 24845553 24438588 25244985 20566863 24495485 22516182 25329095 26108605 28004739 17994087 25069045 15632085 9425132 12530964 24755649 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22751099 23594743 10585570 11100729 25463417
EMBL
BABH01001586
BABH01001587
BABH01001588
RSAL01000295
RVE42832.1
ODYU01002872
+ More
ODYU01002873 SOQ40891.1 SOQ40893.1 NWSH01000842 PCG73890.1 KQ459606 KPI90936.1 AK401351 BAM17973.1 KQ460556 KPJ13994.1 AGBW02012556 OWR44831.1 NEVH01013241 PNF29570.1 ADMH02000906 ETN64702.1 AJVK01059319 CH477238 EAT46528.1 UFQT01000011 SSX17592.1 GDHF01033893 GDHF01016277 GDHF01008994 JAI18421.1 JAI36037.1 JAI43320.1 GBXI01013973 GBXI01005251 JAD00319.1 JAD09041.1 GAKP01000440 JAC58512.1 KB630786 ERL83870.1 APCN01001416 JXUM01061469 JXUM01061470 KQ562151 KXJ76561.1 AXCM01004569 KQ971338 EFA01689.2 AAAB01008823 EAA05540.4 KK852478 KDR23028.1 ATLV01025458 KE525389 KFB52403.1 GGMR01013275 MBY25894.1 GDHF01007062 JAI45252.1 GEMB01001958 JAS01219.1 GECU01034960 JAS72746.1 AAZO01007376 DS232497 EDS42647.1 DS235882 EEB20152.1 GECZ01029758 JAS40011.1 GEBQ01012677 JAT27300.1 CCAG010014160 GAMC01015674 GAMC01015667 JAB90888.1 GAMC01015673 GAMC01015668 JAB90887.1 ABLF02037278 GGMS01004273 MBY73476.1 BT128007 AEE62969.1 GFXV01007880 MBW19685.1 KB632126 ERL88955.1 JRES01000955 KNC26803.1 GEZM01083945 JAV60920.1 OUUW01000025 SPP89614.1 CH479209 EDW32701.1 CH963851 EDW75069.2 JH431265 CH379064 EAL31606.2 CU458990 CAM73251.1 AF006488 BC076414 RJVU01055763 ROK74152.1 FR913631 CDQ92677.1 CM000162 EDX02035.1 AE014298 BT122182 AAF47984.1 ADE58552.1 CH916376 EDV95388.1 CH954180 EDV46111.1 CH940655 EDW66292.2 JH883261 ELR46918.1 AL935335 CU458991 CAM73252.1 AF146429
ODYU01002873 SOQ40891.1 SOQ40893.1 NWSH01000842 PCG73890.1 KQ459606 KPI90936.1 AK401351 BAM17973.1 KQ460556 KPJ13994.1 AGBW02012556 OWR44831.1 NEVH01013241 PNF29570.1 ADMH02000906 ETN64702.1 AJVK01059319 CH477238 EAT46528.1 UFQT01000011 SSX17592.1 GDHF01033893 GDHF01016277 GDHF01008994 JAI18421.1 JAI36037.1 JAI43320.1 GBXI01013973 GBXI01005251 JAD00319.1 JAD09041.1 GAKP01000440 JAC58512.1 KB630786 ERL83870.1 APCN01001416 JXUM01061469 JXUM01061470 KQ562151 KXJ76561.1 AXCM01004569 KQ971338 EFA01689.2 AAAB01008823 EAA05540.4 KK852478 KDR23028.1 ATLV01025458 KE525389 KFB52403.1 GGMR01013275 MBY25894.1 GDHF01007062 JAI45252.1 GEMB01001958 JAS01219.1 GECU01034960 JAS72746.1 AAZO01007376 DS232497 EDS42647.1 DS235882 EEB20152.1 GECZ01029758 JAS40011.1 GEBQ01012677 JAT27300.1 CCAG010014160 GAMC01015674 GAMC01015667 JAB90888.1 GAMC01015673 GAMC01015668 JAB90887.1 ABLF02037278 GGMS01004273 MBY73476.1 BT128007 AEE62969.1 GFXV01007880 MBW19685.1 KB632126 ERL88955.1 JRES01000955 KNC26803.1 GEZM01083945 JAV60920.1 OUUW01000025 SPP89614.1 CH479209 EDW32701.1 CH963851 EDW75069.2 JH431265 CH379064 EAL31606.2 CU458990 CAM73251.1 AF006488 BC076414 RJVU01055763 ROK74152.1 FR913631 CDQ92677.1 CM000162 EDX02035.1 AE014298 BT122182 AAF47984.1 ADE58552.1 CH916376 EDV95388.1 CH954180 EDV46111.1 CH940655 EDW66292.2 JH883261 ELR46918.1 AL935335 CU458991 CAM73252.1 AF146429
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000053240
UP000007151
+ More
UP000235965 UP000095300 UP000075884 UP000000673 UP000092462 UP000095301 UP000008820 UP000075920 UP000075900 UP000076407 UP000075902 UP000075881 UP000075882 UP000030742 UP000075903 UP000075840 UP000075885 UP000069940 UP000249989 UP000069272 UP000075883 UP000007266 UP000007062 UP000027135 UP000030765 UP000075880 UP000076408 UP000075901 UP000002320 UP000009046 UP000092445 UP000092444 UP000078200 UP000007819 UP000018467 UP000091820 UP000037069 UP000268350 UP000008744 UP000007798 UP000265140 UP000001819 UP000221080 UP000261440 UP000261420 UP000000437 UP000261360 UP000193380 UP000002282 UP000087266 UP000264820 UP000000803 UP000001070 UP000007635 UP000008711 UP000008792 UP000192221 UP000265040 UP000261520
UP000235965 UP000095300 UP000075884 UP000000673 UP000092462 UP000095301 UP000008820 UP000075920 UP000075900 UP000076407 UP000075902 UP000075881 UP000075882 UP000030742 UP000075903 UP000075840 UP000075885 UP000069940 UP000249989 UP000069272 UP000075883 UP000007266 UP000007062 UP000027135 UP000030765 UP000075880 UP000076408 UP000075901 UP000002320 UP000009046 UP000092445 UP000092444 UP000078200 UP000007819 UP000018467 UP000091820 UP000037069 UP000268350 UP000008744 UP000007798 UP000265140 UP000001819 UP000221080 UP000261440 UP000261420 UP000000437 UP000261360 UP000193380 UP000002282 UP000087266 UP000264820 UP000000803 UP000001070 UP000007635 UP000008711 UP000008792 UP000192221 UP000265040 UP000261520
Pfam
Interpro
IPR002049
Laminin_EGF
+ More
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR001881 EGF-like_Ca-bd_dom
IPR001774 DSL
IPR016024 ARM-type_fold
IPR009030 Growth_fac_rcpt_cys_sf
IPR029058 AB_hydrolase
IPR000073 AB_hydrolase_1
IPR013111 EGF_extracell
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR018097 EGF_Ca-bd_CS
IPR011651 Notch_ligand_N
IPR004274 FCP1_dom
IPR036412 HAD-like_sf
IPR023214 HAD_sf
IPR013032 EGF-like_CS
IPR000742 EGF-like_dom
IPR001881 EGF-like_Ca-bd_dom
IPR001774 DSL
IPR016024 ARM-type_fold
IPR009030 Growth_fac_rcpt_cys_sf
IPR029058 AB_hydrolase
IPR000073 AB_hydrolase_1
IPR013111 EGF_extracell
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR018097 EGF_Ca-bd_CS
IPR011651 Notch_ligand_N
IPR004274 FCP1_dom
IPR036412 HAD-like_sf
IPR023214 HAD_sf
Gene 3D
ProteinModelPortal
H9JFU8
A0A3S2NLG9
A0A2H1VJ88
A0A2A4JR51
A0A194PCC7
I4DJ83
+ More
A0A0N0PCG8 A0A212ETQ4 A0A2J7QLV8 A0A1I8PB87 A0A182NC65 W5JJZ3 A0A1B0DG68 A0A1I8MRK7 Q17IN2 A0A336LHW1 A0A1S4F1B8 A0A182WFV9 A0A0K8VB06 A0A0A1WMS8 A0A182RYW0 A0A034WVX6 A0A182WUN4 A0A182U5Y4 A0A182KCR2 A0A182KKC3 U4U1J1 A0A182UQG4 A0A182HVU1 A0A182PJ98 A0A182GGA6 A0A182FKD6 A0A182MDB1 D2A0G9 Q7QGY2 A0A067RJ30 A0A084WQA9 A0A182IWL1 A0A182YKN3 A0A182SV86 A0A2S2P900 A0A0K8W2B8 A0A161MR17 A0A1B6HDI0 A0A1S4N2N7 B0X8Q5 E0W3E6 A0A1B0A1B0 A0A1B6EQ11 A0A1B6LUF6 A0A1B0G9S4 W8B1I0 W8AXK5 A0A1A9VWM8 J9JWS8 A0A2S2Q6W0 J3JXN7 A0A2H8TZD5 U4UGD5 A0A3B1ISZ0 A0A3B1K7P8 W5KPC7 A0A1A9WWW1 A0A0L0C381 A0A1Y1KKZ3 A0A3B0K5Y3 B4H4T9 B4MSI0 A0A3P8YLV6 T1IPW5 Q29I00 A0A2D0QCF8 A0A3B4DQL0 W5KGW2 A0A3P8YLP6 A0A3B4VBM2 Q1LYD2 O57409 A0A3B4WWN9 A0A3B4VBN9 A0A3B4X6T1 A0A3N0Y074 A0A060YM88 B4PYV7 A0A1S3RLH3 A0A3Q3DVC3 A0A1S3QDA7 Q9VZ44 B4JXB2 G3Q9T1 B3NVC3 B4MAN3 L8HSS5 A0A1W4V2L9 A0A2R8QUE3 A4JYS0 Q9IAT6 A0A3Q1K3S9 A0A3Q2XGU1 A0A3B4BGD1
A0A0N0PCG8 A0A212ETQ4 A0A2J7QLV8 A0A1I8PB87 A0A182NC65 W5JJZ3 A0A1B0DG68 A0A1I8MRK7 Q17IN2 A0A336LHW1 A0A1S4F1B8 A0A182WFV9 A0A0K8VB06 A0A0A1WMS8 A0A182RYW0 A0A034WVX6 A0A182WUN4 A0A182U5Y4 A0A182KCR2 A0A182KKC3 U4U1J1 A0A182UQG4 A0A182HVU1 A0A182PJ98 A0A182GGA6 A0A182FKD6 A0A182MDB1 D2A0G9 Q7QGY2 A0A067RJ30 A0A084WQA9 A0A182IWL1 A0A182YKN3 A0A182SV86 A0A2S2P900 A0A0K8W2B8 A0A161MR17 A0A1B6HDI0 A0A1S4N2N7 B0X8Q5 E0W3E6 A0A1B0A1B0 A0A1B6EQ11 A0A1B6LUF6 A0A1B0G9S4 W8B1I0 W8AXK5 A0A1A9VWM8 J9JWS8 A0A2S2Q6W0 J3JXN7 A0A2H8TZD5 U4UGD5 A0A3B1ISZ0 A0A3B1K7P8 W5KPC7 A0A1A9WWW1 A0A0L0C381 A0A1Y1KKZ3 A0A3B0K5Y3 B4H4T9 B4MSI0 A0A3P8YLV6 T1IPW5 Q29I00 A0A2D0QCF8 A0A3B4DQL0 W5KGW2 A0A3P8YLP6 A0A3B4VBM2 Q1LYD2 O57409 A0A3B4WWN9 A0A3B4VBN9 A0A3B4X6T1 A0A3N0Y074 A0A060YM88 B4PYV7 A0A1S3RLH3 A0A3Q3DVC3 A0A1S3QDA7 Q9VZ44 B4JXB2 G3Q9T1 B3NVC3 B4MAN3 L8HSS5 A0A1W4V2L9 A0A2R8QUE3 A4JYS0 Q9IAT6 A0A3Q1K3S9 A0A3Q2XGU1 A0A3B4BGD1
PDB
4XLW
E-value=1.14494e-16,
Score=205
Ontologies
GO
GO:0005509
GO:0016021
GO:0016020
GO:0007154
GO:0016298
GO:0044255
GO:0005112
GO:0005886
GO:0045746
GO:0007275
GO:0007219
GO:0030901
GO:0021536
GO:0030154
GO:0035907
GO:0001757
GO:0002244
GO:0060842
GO:0042802
GO:0061056
GO:0048471
GO:0048514
GO:0021514
GO:0005737
GO:0030165
GO:0048666
GO:0060218
GO:0001756
GO:0005515
GO:0006508
GO:0008237
GO:0050660
GO:0046872
GO:0051537
GO:0003824
GO:0016614
GO:0005506
Topology
Subcellular location
Membrane
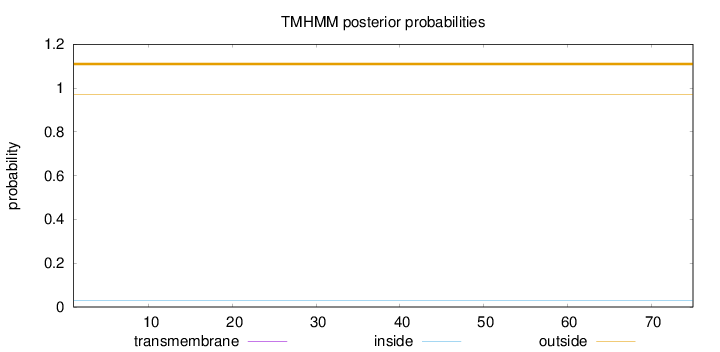
Length:
75
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.02922
outside
1 - 75
Population Genetic Test Statistics
Pi
58.110576
Theta
102.418806
Tajima's D
-1.356658
CLR
180.662552
CSRT
0.07999600019999
Interpretation
Uncertain
