Gene
KWMTBOMO10771
Pre Gene Modal
BGIBMGA008423
Annotation
PREDICTED:_endothelin-converting_enzyme_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.709
Sequence
CDS
ATGGATGCTTGGTCGGCCGGAAATGAACGAGGTCAAGCGGATGCTATGGGGCCCACTGTCACTCGTACCGCAAGTGCTAAGACGAAGACCCCCTCGCCACCACCGCTCAAAAACGGCAAAGATATCCGGGAGTCAGGGTATCTAATAGCTGTTGGATCCAGAACTGCCAGATTCCGTCGAAAACACAAAAAAAGCATGACCCTCATTGTAATTTTAACCCTGATGGCATTTTTATTGATCTTTGCCATCGTAATGACAGTTTTATATGTTCGACAAAAACACAAAAAGCCCGTCCGTTTGTGTGAAACTAAGGAGTGTCTGCGTTCAGCTGCTAGCTTGGCGCTGTCCATGGACCGAACAGCCGATCCTTGCGAAGACTTCAATCAGTATGTCTGTGGCAATTGGGGGGAGGAGCATCCAATGCCCGAAGAATACAGCTCATACGATTGGTTTAGCTACAAGCAAGACAGAGTGCTTGCTTCCATCAAGCTATTTCTCACGAAAAACTCCAGCAAGGATCCCCTGCCGATAAAACAAGCTAGAGATATGTACGCCGCTTGTATGGATGTCGGCAAAATTGACCAACTGGGCCACCAGCCTGTGTACTCCATTTTAGCCTCGATAGGCCTTCCTTCACACCCAACATACCTCAACATCTCAGAAGTAGATTACTCTACATATACATTCGACTGGGTGGGAGTGGTCACAAACATCAAAGTGCTTCTAGGCATGGATATCCTTATTGGATTCGATATATTCTCAGATCCCTTGAACTCGTCCGCTTACCGTCTAGCATTGGGTTCGCCTGAAACAACCAATCCATTTCCAAGCTTGTACGCAATGCAACGTAAATGGGCCAGAAAAGAATTCCCATTGAGGCAACTCCCAAAACAGAAAGAACATAGCGACTTAATGTACCTCACTCATGATGAAGAAACGTACGCCAAAGTGCACAAACTGTACTACGCTGAACTGTTGAAGATTTTTCTTGAAGGAGAAGGTGGAAACCAAACTGCAAGTTTTGACAATGATGTTATCGTGGAAAAGGTCATAGCCGCTGCCGCAACTTATTATGATATGCGGAATGATTTGTACGACATCGAATCTGAGAACGATACCGAAAGTGAAATAGAAAATTACGTTATACCAAGATACACAGCGGACGAACTCCAAGCCTTAACAGATCAAACCGTTGCTCAAAACAATGGAAGCGTAGTCCCTATATGGAAGAAATACGTGGAAGGGGTATTTAATGTTAGTGAAACTAAACTGGATTTTGGTAAAGAGAAAATATTAGTTGCGGACTTAGATTTGAAGTACATGTCGCGTATGGCAGCGTACGTGGCGAAAATTCATCCCGTTGTTCTGGAATTGTATGTTTGGGTTAAGGTTGTAGAAGTTATAGCAATACACACTACCTCGGAACTCCGATCCCTGTACTACGATGCATACAATGATCTGCAAGAAAGTGACACTAAGATCCCCACTAGAGCACATCTATGTACTGAAGCTGTCAATGATATGATGGGCATGGCAGTGGCTTACGGTATCGCCGATCCTGACTTCGTTAATAATTCAAAACCAAAGATTGAAAGGATGATATATGAATTGAAAAATTCCTTAGCACATCTAGTCGGTCAAGTCAAATGGATGGATGACAACACAAAAGTGGCAACATACCAAAAGATTATATTGATGAAGTCTCTAATAGGATTTCCTGATTGGCTAATGGTCGAGGGGAAGTTAGAGGAATATTACAAAGGTATAGAGATCAACAGAGAAACTCACCTGTACAATATGATAAACATAATCCAAGTCAAAATAAAGAACGTGCTAAACAAATTCCGGGAGAATGGCAGTTTGTCTTGGGCTACAGATCCTACTGAAGTTAACGCTTACCACACATTCCAGGATAACACTATAACCGTACCTCTTGTTATATTACAATATCCTTTTTTTGACTTGGGATTGGATTCGTTGAATTACGGCTCACTAGGCACTGTACTTGGACACGAAATAACTCACGGTTTTGATAGTATTGGTCGCAGATACGACAAATTCGGTAACATGGTCCCATGGTGGACCAATAACACAATAGATACGTTTGTCAACATGTCTCGGTGCTTCGTTGATCAATACTCTGCGTACTACCTTCCTGAGATCAATGAACACGTCAGTGGTAAAAACACTTTACCGGAAAATATAGCAGATAATGGGGGAGTCAGAGAAGCCATTGCAGCATTGAAAGAACACCTTAAGAAATTTGGTCCCGAACTCAAGCTACCTGGCTTCGAAGAAATGTCGCCCGAGCAATTATTCTTTCTATCCTATGGAAATCTCTGGTGTGGAACTTCAACAAAAAGCTCGTTAAAGGTCAACCTGAAAGACGAGCACACCCCTCTGCCCTTCAGAGCAAAGGGCGTCCTGCAGAACAGCGAAGAGTTCTCCAGGGCTTTCAAGTGCCGCCCTGGGTCGTCAATGAACCCGAATAAGCGATGTATTATATTTTAA
Protein
MDAWSAGNERGQADAMGPTVTRTASAKTKTPSPPPLKNGKDIRESGYLIAVGSRTARFRRKHKKSMTLIVILTLMAFLLIFAIVMTVLYVRQKHKKPVRLCETKECLRSAASLALSMDRTADPCEDFNQYVCGNWGEEHPMPEEYSSYDWFSYKQDRVLASIKLFLTKNSSKDPLPIKQARDMYAACMDVGKIDQLGHQPVYSILASIGLPSHPTYLNISEVDYSTYTFDWVGVVTNIKVLLGMDILIGFDIFSDPLNSSAYRLALGSPETTNPFPSLYAMQRKWARKEFPLRQLPKQKEHSDLMYLTHDEETYAKVHKLYYAELLKIFLEGEGGNQTASFDNDVIVEKVIAAAATYYDMRNDLYDIESENDTESEIENYVIPRYTADELQALTDQTVAQNNGSVVPIWKKYVEGVFNVSETKLDFGKEKILVADLDLKYMSRMAAYVAKIHPVVLELYVWVKVVEVIAIHTTSELRSLYYDAYNDLQESDTKIPTRAHLCTEAVNDMMGMAVAYGIADPDFVNNSKPKIERMIYELKNSLAHLVGQVKWMDDNTKVATYQKIILMKSLIGFPDWLMVEGKLEEYYKGIEINRETHLYNMINIIQVKIKNVLNKFRENGSLSWATDPTEVNAYHTFQDNTITVPLVILQYPFFDLGLDSLNYGSLGTVLGHEITHGFDSIGRRYDKFGNMVPWWTNNTIDTFVNMSRCFVDQYSAYYLPEINEHVSGKNTLPENIADNGGVREAIAALKEHLKKFGPELKLPGFEEMSPEQLFFLSYGNLWCGTSTKSSLKVNLKDEHTPLPFRAKGVLQNSEEFSRAFKCRPGSSMNPNKRCIIF
Summary
Uniprot
H9JFX6
A0A2A4JR15
A0A2H1VKB7
A0A3S2L0K9
A0A212ETM5
A0A0L7LAG9
+ More
A0A194PD54 A0A182HA66 A0A1L8E2K3 A0A182VEC0 A0A1S4GF43 A0A182LF63 Q7QC43 A0A182HT26 A0A182XWZ4 A0A182R9Y4 A0A182NFH6 A0A182VXA8 A0A084VU62 A0A182TPZ7 A0A182TCQ0 B0X6H4 A0A182P744 A0A182KGI1 B4N8E8 B3MT13 B4QXD5 B4JHG7 B4PRW2 A0A1Y1LBL7 A0A3B0KAF8 Q8IMQ2 A0A182JAC9 A0A1I8MIK5 B4M119 B3P867 B4IC31 B4K9P1 A0A1I8P4T7 A0A1W4U5C3 A0A0L0BKJ5 I5ANE4 A0A0M3QXA6 A0A195BMZ6 A0A0M9A1F9 A0A087ZVK7 A0A2J7RCK5 A0A1B0G3N3 A0A154P2K6 A0A1A9VZB7 A0A0L7RAA7 A0A1W4WHQ1 A0A158P0I7 A0A1W4W7C1 Q16WF8 A0A1S4FK06 A0A1Q3FQW0 K7IM52 F4WIG5 A0A1Q3FPJ5 A0A1B0B325 Q16YF9 A0A1A9YGR0 D2A0F3 A0A1J1HU77 B0X6H3 A0A1W4WIQ8 A0A023EXT2 A0A195F2J0 A0A182HDF7 A0A023EW22 J9HSZ2 A0A182HDF8 A0A2A3ESD8 A0A232EUP6 U4UDU0 F4WIG6 A0A195BLG0 A0A2J7R1I2 A0A023EUQ9 A0A2J7R1H9 A0A1B6DM23 A0A1B6KZB7 A0A158NWD7 A0A151HZV3 F4W4P9 A0A088ANV7 A0A195EVW1 A0A151WUV5 A0A195E2Y7 E2ALK7 A0A2A3EQM5 A0A0C9QE15 A0A0C9PKK5 E2BED8 A0A069DW50
A0A194PD54 A0A182HA66 A0A1L8E2K3 A0A182VEC0 A0A1S4GF43 A0A182LF63 Q7QC43 A0A182HT26 A0A182XWZ4 A0A182R9Y4 A0A182NFH6 A0A182VXA8 A0A084VU62 A0A182TPZ7 A0A182TCQ0 B0X6H4 A0A182P744 A0A182KGI1 B4N8E8 B3MT13 B4QXD5 B4JHG7 B4PRW2 A0A1Y1LBL7 A0A3B0KAF8 Q8IMQ2 A0A182JAC9 A0A1I8MIK5 B4M119 B3P867 B4IC31 B4K9P1 A0A1I8P4T7 A0A1W4U5C3 A0A0L0BKJ5 I5ANE4 A0A0M3QXA6 A0A195BMZ6 A0A0M9A1F9 A0A087ZVK7 A0A2J7RCK5 A0A1B0G3N3 A0A154P2K6 A0A1A9VZB7 A0A0L7RAA7 A0A1W4WHQ1 A0A158P0I7 A0A1W4W7C1 Q16WF8 A0A1S4FK06 A0A1Q3FQW0 K7IM52 F4WIG5 A0A1Q3FPJ5 A0A1B0B325 Q16YF9 A0A1A9YGR0 D2A0F3 A0A1J1HU77 B0X6H3 A0A1W4WIQ8 A0A023EXT2 A0A195F2J0 A0A182HDF7 A0A023EW22 J9HSZ2 A0A182HDF8 A0A2A3ESD8 A0A232EUP6 U4UDU0 F4WIG6 A0A195BLG0 A0A2J7R1I2 A0A023EUQ9 A0A2J7R1H9 A0A1B6DM23 A0A1B6KZB7 A0A158NWD7 A0A151HZV3 F4W4P9 A0A088ANV7 A0A195EVW1 A0A151WUV5 A0A195E2Y7 E2ALK7 A0A2A3EQM5 A0A0C9QE15 A0A0C9PKK5 E2BED8 A0A069DW50
Pubmed
19121390
22118469
26227816
26354079
26483478
12364791
+ More
20966253 25244985 24438588 17994087 17550304 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 26108605 15632085 21347285 17510324 20075255 21719571 18362917 19820115 24945155 28648823 23537049 20798317 26334808
20966253 25244985 24438588 17994087 17550304 28004739 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25315136 26108605 15632085 21347285 17510324 20075255 21719571 18362917 19820115 24945155 28648823 23537049 20798317 26334808
EMBL
BABH01001585
NWSH01000842
PCG73893.1
ODYU01002872
SOQ40892.1
RSAL01000295
+ More
RVE42834.1 AGBW02012556 OWR44829.1 JTDY01001984 KOB72405.1 KQ459606 KPI90938.1 JXUM01122146 KQ566573 KXJ70021.1 GFDF01001282 JAV12802.1 AAAB01008859 EAA08074.4 APCN01000403 ATLV01016645 KE525099 KFB41506.1 DS232413 EDS41457.1 CH964232 EDW81399.2 CH902623 EDV30403.2 CM000364 EDX14621.1 CH916369 EDV92794.1 CM000160 EDW98555.1 GEZM01062408 JAV69740.1 OUUW01000005 SPP80548.1 AE014297 AAN14386.2 CH940650 EDW67430.1 CH954182 EDV53471.1 CH480828 EDW45189.1 CH933806 EDW14516.1 JRES01001712 KNC20605.1 CM000070 EIM52479.2 CP012526 ALC45593.1 KQ976440 KYM86537.1 KQ435763 KOX75390.1 NEVH01005885 PNF38574.1 CCAG010005690 KQ434804 KZC06063.1 KQ414618 KOC67822.1 ADTU01005088 ADTU01005089 ADTU01005090 ADTU01005091 ADTU01005092 CH477568 EAT38904.1 GFDL01005034 JAV30011.1 GL888175 EGI66096.1 GFDL01005558 JAV29487.1 JXJN01007763 JXJN01007764 JXJN01007765 CH477516 EAT39697.1 KQ971338 EFA02504.2 CVRI01000021 CRK91555.1 EDS41456.1 GAPW01000274 JAC13324.1 KQ981856 KYN34606.1 JXUM01129462 KQ567449 KXJ69421.1 GAPW01000271 JAC13327.1 EJY57758.1 JXUM01129464 KXJ69422.1 KZ288193 PBC34126.1 NNAY01002112 OXU22046.1 KB632278 ERL91222.1 EGI66097.1 KYM86538.1 NEVH01008206 PNF34684.1 GAPW01000433 JAC13165.1 PNF34683.1 GEDC01010569 JAS26729.1 GEBQ01023217 JAT16760.1 ADTU01027908 ADTU01027909 KQ976657 KYM78626.1 GL887547 EGI70815.1 KQ981954 KYN32037.1 KQ982718 KYQ51710.1 KQ979701 KYN19530.1 GL440609 EFN65709.1 PBC34065.1 GBYB01001559 GBYB01001561 JAG71326.1 JAG71328.1 GBYB01001558 JAG71325.1 GL447768 EFN85983.1 GBGD01000596 JAC88293.1
RVE42834.1 AGBW02012556 OWR44829.1 JTDY01001984 KOB72405.1 KQ459606 KPI90938.1 JXUM01122146 KQ566573 KXJ70021.1 GFDF01001282 JAV12802.1 AAAB01008859 EAA08074.4 APCN01000403 ATLV01016645 KE525099 KFB41506.1 DS232413 EDS41457.1 CH964232 EDW81399.2 CH902623 EDV30403.2 CM000364 EDX14621.1 CH916369 EDV92794.1 CM000160 EDW98555.1 GEZM01062408 JAV69740.1 OUUW01000005 SPP80548.1 AE014297 AAN14386.2 CH940650 EDW67430.1 CH954182 EDV53471.1 CH480828 EDW45189.1 CH933806 EDW14516.1 JRES01001712 KNC20605.1 CM000070 EIM52479.2 CP012526 ALC45593.1 KQ976440 KYM86537.1 KQ435763 KOX75390.1 NEVH01005885 PNF38574.1 CCAG010005690 KQ434804 KZC06063.1 KQ414618 KOC67822.1 ADTU01005088 ADTU01005089 ADTU01005090 ADTU01005091 ADTU01005092 CH477568 EAT38904.1 GFDL01005034 JAV30011.1 GL888175 EGI66096.1 GFDL01005558 JAV29487.1 JXJN01007763 JXJN01007764 JXJN01007765 CH477516 EAT39697.1 KQ971338 EFA02504.2 CVRI01000021 CRK91555.1 EDS41456.1 GAPW01000274 JAC13324.1 KQ981856 KYN34606.1 JXUM01129462 KQ567449 KXJ69421.1 GAPW01000271 JAC13327.1 EJY57758.1 JXUM01129464 KXJ69422.1 KZ288193 PBC34126.1 NNAY01002112 OXU22046.1 KB632278 ERL91222.1 EGI66097.1 KYM86538.1 NEVH01008206 PNF34684.1 GAPW01000433 JAC13165.1 PNF34683.1 GEDC01010569 JAS26729.1 GEBQ01023217 JAT16760.1 ADTU01027908 ADTU01027909 KQ976657 KYM78626.1 GL887547 EGI70815.1 KQ981954 KYN32037.1 KQ982718 KYQ51710.1 KQ979701 KYN19530.1 GL440609 EFN65709.1 PBC34065.1 GBYB01001559 GBYB01001561 JAG71326.1 JAG71328.1 GBYB01001558 JAG71325.1 GL447768 EFN85983.1 GBGD01000596 JAC88293.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000007151
UP000037510
UP000053268
+ More
UP000069940 UP000249989 UP000075903 UP000075882 UP000007062 UP000075840 UP000076408 UP000075900 UP000075884 UP000075920 UP000030765 UP000075902 UP000075901 UP000002320 UP000075885 UP000075881 UP000007798 UP000007801 UP000000304 UP000001070 UP000002282 UP000268350 UP000000803 UP000075880 UP000095301 UP000008792 UP000008711 UP000001292 UP000009192 UP000095300 UP000192221 UP000037069 UP000001819 UP000092553 UP000078540 UP000053105 UP000005203 UP000235965 UP000092444 UP000076502 UP000091820 UP000053825 UP000192223 UP000005205 UP000008820 UP000002358 UP000007755 UP000092460 UP000092443 UP000007266 UP000183832 UP000078541 UP000242457 UP000215335 UP000030742 UP000075809 UP000078492 UP000000311 UP000008237
UP000069940 UP000249989 UP000075903 UP000075882 UP000007062 UP000075840 UP000076408 UP000075900 UP000075884 UP000075920 UP000030765 UP000075902 UP000075901 UP000002320 UP000075885 UP000075881 UP000007798 UP000007801 UP000000304 UP000001070 UP000002282 UP000268350 UP000000803 UP000075880 UP000095301 UP000008792 UP000008711 UP000001292 UP000009192 UP000095300 UP000192221 UP000037069 UP000001819 UP000092553 UP000078540 UP000053105 UP000005203 UP000235965 UP000092444 UP000076502 UP000091820 UP000053825 UP000192223 UP000005205 UP000008820 UP000002358 UP000007755 UP000092460 UP000092443 UP000007266 UP000183832 UP000078541 UP000242457 UP000215335 UP000030742 UP000075809 UP000078492 UP000000311 UP000008237
Interpro
SUPFAM
SSF57501
SSF57501
Gene 3D
CDD
ProteinModelPortal
H9JFX6
A0A2A4JR15
A0A2H1VKB7
A0A3S2L0K9
A0A212ETM5
A0A0L7LAG9
+ More
A0A194PD54 A0A182HA66 A0A1L8E2K3 A0A182VEC0 A0A1S4GF43 A0A182LF63 Q7QC43 A0A182HT26 A0A182XWZ4 A0A182R9Y4 A0A182NFH6 A0A182VXA8 A0A084VU62 A0A182TPZ7 A0A182TCQ0 B0X6H4 A0A182P744 A0A182KGI1 B4N8E8 B3MT13 B4QXD5 B4JHG7 B4PRW2 A0A1Y1LBL7 A0A3B0KAF8 Q8IMQ2 A0A182JAC9 A0A1I8MIK5 B4M119 B3P867 B4IC31 B4K9P1 A0A1I8P4T7 A0A1W4U5C3 A0A0L0BKJ5 I5ANE4 A0A0M3QXA6 A0A195BMZ6 A0A0M9A1F9 A0A087ZVK7 A0A2J7RCK5 A0A1B0G3N3 A0A154P2K6 A0A1A9VZB7 A0A0L7RAA7 A0A1W4WHQ1 A0A158P0I7 A0A1W4W7C1 Q16WF8 A0A1S4FK06 A0A1Q3FQW0 K7IM52 F4WIG5 A0A1Q3FPJ5 A0A1B0B325 Q16YF9 A0A1A9YGR0 D2A0F3 A0A1J1HU77 B0X6H3 A0A1W4WIQ8 A0A023EXT2 A0A195F2J0 A0A182HDF7 A0A023EW22 J9HSZ2 A0A182HDF8 A0A2A3ESD8 A0A232EUP6 U4UDU0 F4WIG6 A0A195BLG0 A0A2J7R1I2 A0A023EUQ9 A0A2J7R1H9 A0A1B6DM23 A0A1B6KZB7 A0A158NWD7 A0A151HZV3 F4W4P9 A0A088ANV7 A0A195EVW1 A0A151WUV5 A0A195E2Y7 E2ALK7 A0A2A3EQM5 A0A0C9QE15 A0A0C9PKK5 E2BED8 A0A069DW50
A0A194PD54 A0A182HA66 A0A1L8E2K3 A0A182VEC0 A0A1S4GF43 A0A182LF63 Q7QC43 A0A182HT26 A0A182XWZ4 A0A182R9Y4 A0A182NFH6 A0A182VXA8 A0A084VU62 A0A182TPZ7 A0A182TCQ0 B0X6H4 A0A182P744 A0A182KGI1 B4N8E8 B3MT13 B4QXD5 B4JHG7 B4PRW2 A0A1Y1LBL7 A0A3B0KAF8 Q8IMQ2 A0A182JAC9 A0A1I8MIK5 B4M119 B3P867 B4IC31 B4K9P1 A0A1I8P4T7 A0A1W4U5C3 A0A0L0BKJ5 I5ANE4 A0A0M3QXA6 A0A195BMZ6 A0A0M9A1F9 A0A087ZVK7 A0A2J7RCK5 A0A1B0G3N3 A0A154P2K6 A0A1A9VZB7 A0A0L7RAA7 A0A1W4WHQ1 A0A158P0I7 A0A1W4W7C1 Q16WF8 A0A1S4FK06 A0A1Q3FQW0 K7IM52 F4WIG5 A0A1Q3FPJ5 A0A1B0B325 Q16YF9 A0A1A9YGR0 D2A0F3 A0A1J1HU77 B0X6H3 A0A1W4WIQ8 A0A023EXT2 A0A195F2J0 A0A182HDF7 A0A023EW22 J9HSZ2 A0A182HDF8 A0A2A3ESD8 A0A232EUP6 U4UDU0 F4WIG6 A0A195BLG0 A0A2J7R1I2 A0A023EUQ9 A0A2J7R1H9 A0A1B6DM23 A0A1B6KZB7 A0A158NWD7 A0A151HZV3 F4W4P9 A0A088ANV7 A0A195EVW1 A0A151WUV5 A0A195E2Y7 E2ALK7 A0A2A3EQM5 A0A0C9QE15 A0A0C9PKK5 E2BED8 A0A069DW50
PDB
5V48
E-value=5.41375e-75,
Score=718
Ontologies
GO
PANTHER
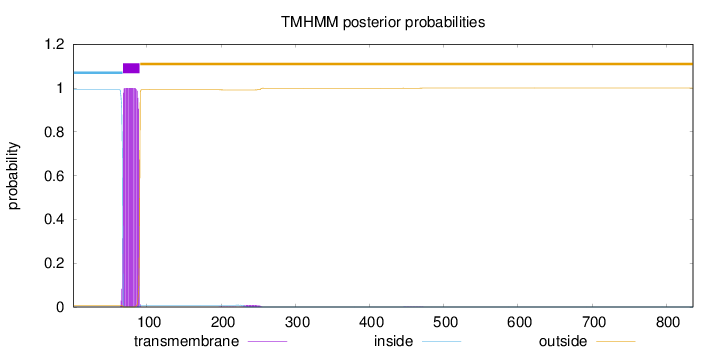
Topology
Length:
836
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.14401
Exp number, first 60 AAs:
0.00032
Total prob of N-in:
0.99218
inside
1 - 67
TMhelix
68 - 90
outside
91 - 836
Population Genetic Test Statistics
Pi
309.356184
Theta
185.761449
Tajima's D
1.925006
CLR
0.22891
CSRT
0.874106294685266
Interpretation
Uncertain
