Pre Gene Modal
BGIBMGA008399
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_symplekin_[Bombyx_mori]
Full name
Symplekin
Location in the cell
Nuclear Reliability : 2.796
Sequence
CDS
ATGTCGAATAAACACGGACACACGTCTAATACTACAGTACAAAATCAGCTAATACGATGGATAAACGATACTGGAATGGCGGAAGGAAACAAAAAAGCAGGCTTATTGAGAAAAGTCATAGAAGTTCTGCTACATCAAGGCTCGCAAATGATTCCAGTATACATGGAGAATATACTAAGCTACATCAGCGATAAAAATACAGACGTAAAGAAGCAGGTCGCGTATTTTGTTGAAGAGTTGAGTAAAAGTCACCCTGAACTTCTTCCCAAGATTGTCGCTCAGTTGCGTCTCCTACTCCTTGACCCAGTCATTGCGGTGCAGAAGAGGGCCATCCAAGCAGCCAGTATACTGTACCGTAATACGTTAATGTGGATCTGTAAAGGTGACGCTGAAGTATCGGAAATGAAACATGTATGGGAACATTTAACTGAATTAAAGTTGATGGTTTTAAACATGATCGACAGTGAGAATGAAGGTATCAGAACGCATTCCATTAAGTTCCTGGAAGAGGTTGTGCTCCGGCAAAGTCCCGGTGACATTGTTGATGCTGAATCCAGCTTGGATAGTCTACCATCAGATGTTCCTTTTATCAATAGAAAAGCTCTCGAAGAGGAATCTGACCATATCTTCAAACTACTGGTCAAATTTCATAATTCACAGCACATATCAAGTGTAAACCTGATGGCTTGTATGACAACGCTATGTTCTTTAGCAAAATTCCGTCCTAAATACTTGCCCCGAGTCATACAAGCACTAGGCGATCTTCACACGACACTACCTCCAACTCTATCGCAGTCTCAAGTCAATTCAGTCAGGAAACATCTCAAAATGCAGATATTGAATTTAATAAAACAATCAACTTCTAATGAGATGATGCCGCAGTTGACACAATTGCTAATTGATATCGGAATGACAGCACAAGAGATTAATAAGGCGGTACCGAAAGAGAAAAGAAACAAACGCTTAGGGGAAATTAGGAATGGGGATACACCATCTAAACGGTTCAGAATCGATTCACCTCAAAGTAATAGTCAAGGTAGCGACAGTAACAGCCGTAGTGAATTCATGTATGACGATGACAGCCAGTCAAGCATCACGAAGCCTTCGGCAACAGAAGACAGTATCCTCGAAGGCTTAAATTCTGTGGACAATGTTGTTAATCTTGTCATGAAAACATTGCAGAATTGTCTACCAGATAACATGCCGCCCTCGTTCACAAATGATTACAAGCCAATACCTAACTCTGGTAGCAAAGTACAAAAGAAGAACTTGGCTAAAATGTTGATGAGCTTAATAAGAGGTGAATCAGAAAATACACAAATAGTTACACAATCAATGGAGGTAGATACACAACCTAACCAGACAGTATTACAACAGAGCTTATCCGATATAGCTGCGAAAATACCGTTAATAAGGGAAGATGATGAGAAAACTACCCTCAAAAATGCTGTAGCTAAATTACAAGAGTCAACAAAAGACAGGCAGATAGAGAGTGCGGTTTCTAAACTAATGGAAGAAACGAGACAGGAACATTTGAAGGAGGAAGAGAGGAAAGCAAAGGAGAAAGAGAAACCGGTTCCTCCACCTACTCCGACGGTGCCTAAACTGAAACAGAAAGTGAAGCTATTGAAACTACAGGAAATAACACGGCCCATCCCAAAAGAAATAAAGCAAAAGTTGATACTGCAGGCCGTTGAGAGGATTTTAAATGCGGAAAAAGACGCCGCTATCGGTGGAGCCGAACAGATCCGGGCCAAATTCATCACGATATTCGCTTCGAGTTACACGCCGGAAACGCGCGATCTAGTACTGAATTACATTTTAGACAATCCCGCCGAGCGCATCAACCTCGCTTTGAAGTGGTTATATGAAGAATACGCATTCATGCAGGGCTTTAACCGACATCCGGTGACCCTACAACCGAAACTGCACGAGAAGCACGATCAAAACTACAACCAGTTGCTCTGTGCTCTGGTCACCCAGATATCCGAGAGAGGCGACCCTGTCATGGAAGGATGCAAAGATGTTTTACTAAGGAAGATATATTTGGAGAGTCCGGTGATCACGGACGAGGCCCTGGATTATTTGAAGCACTTGGTCACTGAAGATCAGGTGGCTCTGCTGGCTTTGGAGTTGCTAGAGGAATTGTGTTTGATGAGACCACCTCGGGCACACAAATATGTGACATCGTTGGTCTGTCATGTTTTGAGCGAAAATGAGGAAATCCGCGATATAGCTTTGAAAGCTACCATCAAAGTTTACAAACGTGGCTCAGAAGCTGTTAAGAAAGTCATCGAGAAACACGCTCTCCTGTACTTAGGGTTCATATCGTTGAATCATCCGCCCCCTGAACTTTCAAGCACCAGAACTAGGATCATGTGGAATGAGAATTTACATAAGCTGTGCTTGAATTTAGTTGTGGCGCTCTTCCCAGAAAAAGAAGGACTACTGATGGAAATAGCAAGGATATACGGCAGCTCCGGTGCAGAGGCGAAGCGCGTTGTACTGCGAACCATCGAGGGTCCGATCCGGGCGATGGTGTCCAAGGACCTGCCCGACAACGAGTTCGACGAGGACGACGCCACGCTGCGCCCCGCCTTCAGGGAACTGCTCGAGAACTGTCCCCGAGGAGCAGAGACTCTGCTCACCAGACTAGTGCATATACTGACCGAGAAAGTGCCTCCAACAGCCGAACTAGTGTCGAAAGTACGAGACATATACGCCACCAGAGTGTCCGACGTTAGATTCTTGATCCCCGTGCTCACCGGACTGTCAAAGAGAGAGATCGTGGCAGCCCTACCGAAACTAATCAAGCTCAACCCTGCCGTCGTGAAGGAGGTGTTCAACAAGCTGTTGGGAATCAACGGCACGTACGACGAGCAGTCGCCCGGTAGGTGGCGCCACCGACGCGGGTCTGAGAGAGAGAGAGAAAGAGAGAGCATACTTTATTGCTCACCAAAACACTATTAA
Protein
MSNKHGHTSNTTVQNQLIRWINDTGMAEGNKKAGLLRKVIEVLLHQGSQMIPVYMENILSYISDKNTDVKKQVAYFVEELSKSHPELLPKIVAQLRLLLLDPVIAVQKRAIQAASILYRNTLMWICKGDAEVSEMKHVWEHLTELKLMVLNMIDSENEGIRTHSIKFLEEVVLRQSPGDIVDAESSLDSLPSDVPFINRKALEEESDHIFKLLVKFHNSQHISSVNLMACMTTLCSLAKFRPKYLPRVIQALGDLHTTLPPTLSQSQVNSVRKHLKMQILNLIKQSTSNEMMPQLTQLLIDIGMTAQEINKAVPKEKRNKRLGEIRNGDTPSKRFRIDSPQSNSQGSDSNSRSEFMYDDDSQSSITKPSATEDSILEGLNSVDNVVNLVMKTLQNCLPDNMPPSFTNDYKPIPNSGSKVQKKNLAKMLMSLIRGESENTQIVTQSMEVDTQPNQTVLQQSLSDIAAKIPLIREDDEKTTLKNAVAKLQESTKDRQIESAVSKLMEETRQEHLKEEERKAKEKEKPVPPPTPTVPKLKQKVKLLKLQEITRPIPKEIKQKLILQAVERILNAEKDAAIGGAEQIRAKFITIFASSYTPETRDLVLNYILDNPAERINLALKWLYEEYAFMQGFNRHPVTLQPKLHEKHDQNYNQLLCALVTQISERGDPVMEGCKDVLLRKIYLESPVITDEALDYLKHLVTEDQVALLALELLEELCLMRPPRAHKYVTSLVCHVLSENEEIRDIALKATIKVYKRGSEAVKKVIEKHALLYLGFISLNHPPPELSSTRTRIMWNENLHKLCLNLVVALFPEKEGLLMEIARIYGSSGAEAKRVVLRTIEGPIRAMVSKDLPDNEFDEDDATLRPAFRELLENCPRGAETLLTRLVHILTEKVPPTAELVSKVRDIYATRVSDVRFLIPVLTGLSKREIVAALPKLIKLNPAVVKEVFNKLLGINGTYDEQSPGRWRHRRGSERERERESILYCSPKHY
Summary
Description
Component of a protein complex required for cotranscriptional processing of 3'-ends of polyadenylated and histone pre-mRNA.
Subunit
Interacts with Cpsf73 and Cpsf100 forming a core cleavage factor required for both polyadenylated and histone mRNA processing. Interacts with Slbp and Lsm11.
Similarity
Belongs to the Symplekin family.
Keywords
3D-structure
Coiled coil
Complete proteome
mRNA processing
Nucleus
Reference proteome
Repeat
RNA-binding
Feature
chain Symplekin
Uniprot
H9JFV2
A0A2A4JBI5
A0A2H1VCZ9
A0A212FCK9
A0A0N1IP64
A0A1E1WJQ7
+ More
A0A212F7B0 A0A194PD64 A0A0L7L8R9 A0A2J7QFW6 E0VQ07 A0A151JQH5 F4W5L8 K7IU32 A0A182GJ60 Q17I55 A0A151I253 A0A146L2G3 A0A1B0ESZ5 A0A1W4UJU2 B0WVE7 A0A2R5L7R2 B4QXJ2 A0A084W6K3 B4PUX6 B4I467 A0A0A1WZ23 A0A1Q3F1T5 B3P2J3 A0A1Q3F080 Q8MSU4 Q7QCG6 A0A182LEI4 A0A182TL26 A0A182HSH7 A0A182USV9 B3LWE7 A0A0N8DVW1 B4G491 A0A0P5QXE2 A0A0P5KVQ8 A0A0P6FCV8 A0A182IR14 A0A0T6AXR2 A0A0P6EDG3 B4NK95 A0A0P5GAJ4 Q297L3 A0A0P6IRX1 A0A0P4YLI3 B4LYQ1 A0A0P6FXH0 A0A0P5Z2Y0 A0A0P6IC67 A0A034VV96 A0A1E1XRG5 A0A164YWS4 A0A0P5K921 E9GTR9 A0A023GMD5 A0A182XAN7 W8BBF0 A0A3B0JRY1 A0A0P6B049 A0A1I8MGP9 A0A1B0GEA9 A0A1I8PAP9 A0A1A9Z6Y3 A0A1A9YL74 A0A0L0CD71 A0A1E1X9R8 A0A1A9X1T0 A0A0P5NTQ6 A0A0N8D301 A0A1A9US56 A0A0A9Y5G0 A0A0P5LX38 A0A1B0C585 A0A0P5XVC4 L9L838 A0A293N1G3
A0A212F7B0 A0A194PD64 A0A0L7L8R9 A0A2J7QFW6 E0VQ07 A0A151JQH5 F4W5L8 K7IU32 A0A182GJ60 Q17I55 A0A151I253 A0A146L2G3 A0A1B0ESZ5 A0A1W4UJU2 B0WVE7 A0A2R5L7R2 B4QXJ2 A0A084W6K3 B4PUX6 B4I467 A0A0A1WZ23 A0A1Q3F1T5 B3P2J3 A0A1Q3F080 Q8MSU4 Q7QCG6 A0A182LEI4 A0A182TL26 A0A182HSH7 A0A182USV9 B3LWE7 A0A0N8DVW1 B4G491 A0A0P5QXE2 A0A0P5KVQ8 A0A0P6FCV8 A0A182IR14 A0A0T6AXR2 A0A0P6EDG3 B4NK95 A0A0P5GAJ4 Q297L3 A0A0P6IRX1 A0A0P4YLI3 B4LYQ1 A0A0P6FXH0 A0A0P5Z2Y0 A0A0P6IC67 A0A034VV96 A0A1E1XRG5 A0A164YWS4 A0A0P5K921 E9GTR9 A0A023GMD5 A0A182XAN7 W8BBF0 A0A3B0JRY1 A0A0P6B049 A0A1I8MGP9 A0A1B0GEA9 A0A1I8PAP9 A0A1A9Z6Y3 A0A1A9YL74 A0A0L0CD71 A0A1E1X9R8 A0A1A9X1T0 A0A0P5NTQ6 A0A0N8D301 A0A1A9US56 A0A0A9Y5G0 A0A0P5LX38 A0A1B0C585 A0A0P5XVC4 L9L838 A0A293N1G3
Pubmed
19121390
22118469
26354079
26227816
20566863
21719571
+ More
20075255 26483478 17510324 26823975 17994087 24438588 17550304 25830018 10731132 12537572 12537569 18042462 19450530 19576221 12364791 14747013 17210077 20966253 15632085 25348373 29209593 21292972 24495485 25315136 26108605 28503490 25401762 23385571
20075255 26483478 17510324 26823975 17994087 24438588 17550304 25830018 10731132 12537572 12537569 18042462 19450530 19576221 12364791 14747013 17210077 20966253 15632085 25348373 29209593 21292972 24495485 25315136 26108605 28503490 25401762 23385571
EMBL
BABH01001574
BABH01001575
BABH01001576
BABH01001577
NWSH01002143
PCG69058.1
+ More
ODYU01001878 SOQ38697.1 AGBW02009168 OWR51479.1 KQ460556 KPJ13980.1 GDQN01003839 JAT87215.1 AGBW02009901 OWR49608.1 KQ459606 KPI90948.1 JTDY01002196 KOB71902.1 NEVH01014841 PNF27475.1 DS235387 EEB15463.1 KQ978642 KYN29485.1 GL887655 EGI70515.1 AAZX01016310 JXUM01069354 KQ562548 KXJ75644.1 CH477242 EAT46374.1 KQ976543 KYM80966.1 GDHC01015888 JAQ02741.1 AJWK01005336 AJWK01005337 AJWK01005338 AJWK01005339 AJWK01005340 DS232125 EDS35552.1 GGLE01001393 MBY05519.1 CM000364 EDX11778.1 ATLV01020836 KE525308 KFB45847.1 CM000160 EDW97783.1 CH480821 EDW55010.1 GBXI01010624 JAD03668.1 GFDL01013546 JAV21499.1 CH954181 EDV47943.1 GFDL01014089 JAV20956.1 AE014297 AY118592 AAF51962.2 AAM49961.1 AAAB01008859 EAA08083.5 APCN01000279 CH902617 EDV43780.1 GDIQ01086780 JAN07957.1 CH479179 EDW25129.1 GDIQ01146616 GDIQ01111917 GDIQ01110927 GDIQ01108126 GDIQ01047903 GDIQ01044298 JAL40799.1 GDIQ01178601 JAK73124.1 GDIQ01061791 JAN32946.1 LJIG01022555 KRT79951.1 GDIQ01064943 JAN29794.1 CH964272 EDW84025.1 GDIQ01265369 JAJ86355.1 CM000070 EAL28192.1 GDIQ01006568 JAN88169.1 GDIP01227758 JAI95643.1 CH940650 EDW66978.1 GDIQ01052305 JAN42432.1 GDIP01050734 JAM52981.1 GDIQ01032203 JAN62534.1 GAKP01012583 GAKP01012578 GAKP01012575 GAKP01012572 JAC46369.1 GFAA01001529 JAU01906.1 LRGB01000868 KZS15691.1 GDIQ01213538 JAK38187.1 GL732564 EFX77164.1 GBBM01000426 JAC34992.1 GAMC01010553 JAB96002.1 OUUW01000007 SPP83152.1 GDIP01021908 JAM81807.1 CCAG010012181 JRES01000562 KNC30191.1 GFAC01003193 JAT95995.1 GDIQ01148077 JAL03649.1 GDIP01073839 JAM29876.1 GBHO01016200 GBRD01006947 JAG27404.1 JAG58874.1 GDIQ01166845 JAK84880.1 JXJN01025888 GDIP01066776 JAM36939.1 KB320473 ELW71131.1 GFWV01021481 MAA46209.1
ODYU01001878 SOQ38697.1 AGBW02009168 OWR51479.1 KQ460556 KPJ13980.1 GDQN01003839 JAT87215.1 AGBW02009901 OWR49608.1 KQ459606 KPI90948.1 JTDY01002196 KOB71902.1 NEVH01014841 PNF27475.1 DS235387 EEB15463.1 KQ978642 KYN29485.1 GL887655 EGI70515.1 AAZX01016310 JXUM01069354 KQ562548 KXJ75644.1 CH477242 EAT46374.1 KQ976543 KYM80966.1 GDHC01015888 JAQ02741.1 AJWK01005336 AJWK01005337 AJWK01005338 AJWK01005339 AJWK01005340 DS232125 EDS35552.1 GGLE01001393 MBY05519.1 CM000364 EDX11778.1 ATLV01020836 KE525308 KFB45847.1 CM000160 EDW97783.1 CH480821 EDW55010.1 GBXI01010624 JAD03668.1 GFDL01013546 JAV21499.1 CH954181 EDV47943.1 GFDL01014089 JAV20956.1 AE014297 AY118592 AAF51962.2 AAM49961.1 AAAB01008859 EAA08083.5 APCN01000279 CH902617 EDV43780.1 GDIQ01086780 JAN07957.1 CH479179 EDW25129.1 GDIQ01146616 GDIQ01111917 GDIQ01110927 GDIQ01108126 GDIQ01047903 GDIQ01044298 JAL40799.1 GDIQ01178601 JAK73124.1 GDIQ01061791 JAN32946.1 LJIG01022555 KRT79951.1 GDIQ01064943 JAN29794.1 CH964272 EDW84025.1 GDIQ01265369 JAJ86355.1 CM000070 EAL28192.1 GDIQ01006568 JAN88169.1 GDIP01227758 JAI95643.1 CH940650 EDW66978.1 GDIQ01052305 JAN42432.1 GDIP01050734 JAM52981.1 GDIQ01032203 JAN62534.1 GAKP01012583 GAKP01012578 GAKP01012575 GAKP01012572 JAC46369.1 GFAA01001529 JAU01906.1 LRGB01000868 KZS15691.1 GDIQ01213538 JAK38187.1 GL732564 EFX77164.1 GBBM01000426 JAC34992.1 GAMC01010553 JAB96002.1 OUUW01000007 SPP83152.1 GDIP01021908 JAM81807.1 CCAG010012181 JRES01000562 KNC30191.1 GFAC01003193 JAT95995.1 GDIQ01148077 JAL03649.1 GDIP01073839 JAM29876.1 GBHO01016200 GBRD01006947 JAG27404.1 JAG58874.1 GDIQ01166845 JAK84880.1 JXJN01025888 GDIP01066776 JAM36939.1 KB320473 ELW71131.1 GFWV01021481 MAA46209.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000037510
+ More
UP000235965 UP000009046 UP000078492 UP000007755 UP000002358 UP000069940 UP000249989 UP000008820 UP000078540 UP000092461 UP000192221 UP000002320 UP000000304 UP000030765 UP000002282 UP000001292 UP000008711 UP000000803 UP000007062 UP000075882 UP000075902 UP000075840 UP000075903 UP000007801 UP000008744 UP000075880 UP000007798 UP000001819 UP000008792 UP000076858 UP000000305 UP000076407 UP000268350 UP000095301 UP000092444 UP000095300 UP000092445 UP000092443 UP000037069 UP000091820 UP000078200 UP000092460 UP000011518
UP000235965 UP000009046 UP000078492 UP000007755 UP000002358 UP000069940 UP000249989 UP000008820 UP000078540 UP000092461 UP000192221 UP000002320 UP000000304 UP000030765 UP000002282 UP000001292 UP000008711 UP000000803 UP000007062 UP000075882 UP000075902 UP000075840 UP000075903 UP000007801 UP000008744 UP000075880 UP000007798 UP000001819 UP000008792 UP000076858 UP000000305 UP000076407 UP000268350 UP000095301 UP000092444 UP000095300 UP000092445 UP000092443 UP000037069 UP000091820 UP000078200 UP000092460 UP000011518
Interpro
Gene 3D
ProteinModelPortal
H9JFV2
A0A2A4JBI5
A0A2H1VCZ9
A0A212FCK9
A0A0N1IP64
A0A1E1WJQ7
+ More
A0A212F7B0 A0A194PD64 A0A0L7L8R9 A0A2J7QFW6 E0VQ07 A0A151JQH5 F4W5L8 K7IU32 A0A182GJ60 Q17I55 A0A151I253 A0A146L2G3 A0A1B0ESZ5 A0A1W4UJU2 B0WVE7 A0A2R5L7R2 B4QXJ2 A0A084W6K3 B4PUX6 B4I467 A0A0A1WZ23 A0A1Q3F1T5 B3P2J3 A0A1Q3F080 Q8MSU4 Q7QCG6 A0A182LEI4 A0A182TL26 A0A182HSH7 A0A182USV9 B3LWE7 A0A0N8DVW1 B4G491 A0A0P5QXE2 A0A0P5KVQ8 A0A0P6FCV8 A0A182IR14 A0A0T6AXR2 A0A0P6EDG3 B4NK95 A0A0P5GAJ4 Q297L3 A0A0P6IRX1 A0A0P4YLI3 B4LYQ1 A0A0P6FXH0 A0A0P5Z2Y0 A0A0P6IC67 A0A034VV96 A0A1E1XRG5 A0A164YWS4 A0A0P5K921 E9GTR9 A0A023GMD5 A0A182XAN7 W8BBF0 A0A3B0JRY1 A0A0P6B049 A0A1I8MGP9 A0A1B0GEA9 A0A1I8PAP9 A0A1A9Z6Y3 A0A1A9YL74 A0A0L0CD71 A0A1E1X9R8 A0A1A9X1T0 A0A0P5NTQ6 A0A0N8D301 A0A1A9US56 A0A0A9Y5G0 A0A0P5LX38 A0A1B0C585 A0A0P5XVC4 L9L838 A0A293N1G3
A0A212F7B0 A0A194PD64 A0A0L7L8R9 A0A2J7QFW6 E0VQ07 A0A151JQH5 F4W5L8 K7IU32 A0A182GJ60 Q17I55 A0A151I253 A0A146L2G3 A0A1B0ESZ5 A0A1W4UJU2 B0WVE7 A0A2R5L7R2 B4QXJ2 A0A084W6K3 B4PUX6 B4I467 A0A0A1WZ23 A0A1Q3F1T5 B3P2J3 A0A1Q3F080 Q8MSU4 Q7QCG6 A0A182LEI4 A0A182TL26 A0A182HSH7 A0A182USV9 B3LWE7 A0A0N8DVW1 B4G491 A0A0P5QXE2 A0A0P5KVQ8 A0A0P6FCV8 A0A182IR14 A0A0T6AXR2 A0A0P6EDG3 B4NK95 A0A0P5GAJ4 Q297L3 A0A0P6IRX1 A0A0P4YLI3 B4LYQ1 A0A0P6FXH0 A0A0P5Z2Y0 A0A0P6IC67 A0A034VV96 A0A1E1XRG5 A0A164YWS4 A0A0P5K921 E9GTR9 A0A023GMD5 A0A182XAN7 W8BBF0 A0A3B0JRY1 A0A0P6B049 A0A1I8MGP9 A0A1B0GEA9 A0A1I8PAP9 A0A1A9Z6Y3 A0A1A9YL74 A0A0L0CD71 A0A1E1X9R8 A0A1A9X1T0 A0A0P5NTQ6 A0A0N8D301 A0A1A9US56 A0A0A9Y5G0 A0A0P5LX38 A0A1B0C585 A0A0P5XVC4 L9L838 A0A293N1G3
PDB
6NPW
E-value=2.05113e-58,
Score=576
Ontologies
GO
PANTHER
Topology
Subcellular location
Nucleus
Concentrates in the histone locus body. With evidence from 5 publications.
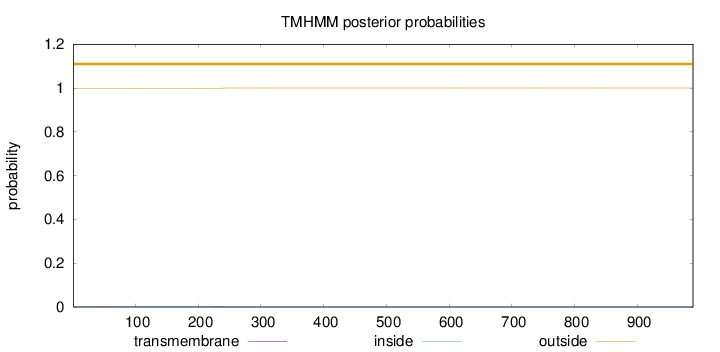
Length:
989
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01956
Exp number, first 60 AAs:
0.00495
Total prob of N-in:
0.00099
outside
1 - 989
Population Genetic Test Statistics
Pi
334.86676
Theta
202.631333
Tajima's D
2.101755
CLR
0.146095
CSRT
0.897955102244888
Interpretation
Uncertain
