Gene
KWMTBOMO10751
Pre Gene Modal
BGIBMGA008401
Annotation
PREDICTED:_alkaline_phosphatase-like_[Bombyx_mori]
Full name
Alkaline phosphatase
Location in the cell
Mitochondrial Reliability : 1.976
Sequence
CDS
ATGACATCGCGACATGCACACTCGACCCATGTTGCAACAGGACTGAGCAAAGTGCAAACTGAAGCAAAGCACCAACATGGGGACGGATCGCGAACGTTGATCCACACGTTGGGCGAGTGGCGTTGGTGCCTATGTTGGGGCCAACGTGTGGAGCCGGCGTTACGTATTGCACTAGTCCACGTGAATCTGACCGTGGCAGTGACTCATGCGTTCATTCACGAGTCAAATAAACGAAAGCGACGCGGTCTAGTCGTTGCCATGTTCCCGCGCATTTTACCTGTGTTCTTTGTTATTTACGCGGCAAAATGTACCTTACGAACAGACCAACAATTCTGGACGGAGCTGGCTGAGAACGAGCTAGCTGAGGCTTTGCAAGTCAAATGGAACCTTGGAGTCGCGAAGAACGTGATCCTCTTCGTCGGAGACGGCATGGGACCTAACACTGTGACGGCGACCAGGATATACAAAGGGGGAGAGAGTCACAGATTGGTTTACGAGAAGTTTCCACATGTTGGACTATTAAAGACATATTCAGCTAACAAAATGGTCCCAGACTCATCGTGTTCCGCCACGGCACTGTTCTGTGGGGTCAAAGCCAATCAAGAGACCATAGGAGTGGACGCTAGTGTGAAGCACAAGGATTGTTCTGCTTCGTTGAGGCCAGAAGCCAGGTTGAAGTCTTTAGCCTCGATAGCTCTGAAAGCTGGGAAGAGTGCTGGTTTTGTGACAACTATGCGAGTCACCCATGCGACCCCGGGTCCCATTTACGCCCACAGTGCTGATCGTAAATGGGAATGCGAAGACAAAATGCCTGCTAGCGCTGCTGCTTGCAAGGATATCGCGAGGCAACTTGTGGAAGACTGGCCCGGAAAAGACTTGCAGGTAATCCTAGGTGGAGGCAGACAGGTTCTTGTATCCAACAGCACAGGTACTAAAGAAGATCCATTAGATACGTGGTCTTGCATCAGAAAGGACGGTAAGGATCTCATAAAAGAATACCAGAAGGACAAACATGACCGCGGCTTGAGATATAGCTATGTATCAAATAACAAGCAGCTTAAAGGATTCGATGTTGACCAAACCGATTATCTTTTGGGAATCTTTGCAAACGGGCATCTAAAATATGAACACGAAAGAAACAAAGGTCCTGAAGGTATGCCTTCGATATCCGATATGGTCGAAGCCGCCATAAAAGTGTTACGGAAAAATAAAAACGGATACTTTCTTATGGTAGAAGGCGGCAATGTGGACATGGCCCACCACCGGGGCTGGGCCAAGCGCGCCATCGACGAGTCCGCGGCCCTCGAGGGCGCCGTGCGGCTCGCGGCTGCGCACACCGACCCCCGGGACACGCTGCTCGTCGTCACCAGCGACCACACGCACACGCTCTCCATCAACGGCTACCCCTACAGGGGCACTGATATATTTGACATAGCCCAAACTTCAAAATACGACAACGTGAACTACACGACGCTGTCATACGGCACCGGAGGCCCGGGCTCCATGAAGTACTCCGTGAAAACCGAACACAACAGGACCGTGGCCGCCAGAGAAGACCCCGGCGTCAACACCGACAGCTTCGACTACCAGCAGATAGCGGCCATCACTCTGGACGAGAACAGCCACGGGGGCGGAGACGTGGTGGTCTACGCGCGAGGTCCCCACGCTCATCTGTTCCACAACGTGCACGAGCAGCATTACGTGTACCACGCGATATCGTACGCCGCCAAAATGGGCGCCTACTCCGGAACCAGCGCCATCTATACCAACATTTCGTTGTTAGCAATCGCCGCCTGGGTTGCCATATGTATAATGAAATAA
Protein
MTSRHAHSTHVATGLSKVQTEAKHQHGDGSRTLIHTLGEWRWCLCWGQRVEPALRIALVHVNLTVAVTHAFIHESNKRKRRGLVVAMFPRILPVFFVIYAAKCTLRTDQQFWTELAENELAEALQVKWNLGVAKNVILFVGDGMGPNTVTATRIYKGGESHRLVYEKFPHVGLLKTYSANKMVPDSSCSATALFCGVKANQETIGVDASVKHKDCSASLRPEARLKSLASIALKAGKSAGFVTTMRVTHATPGPIYAHSADRKWECEDKMPASAAACKDIARQLVEDWPGKDLQVILGGGRQVLVSNSTGTKEDPLDTWSCIRKDGKDLIKEYQKDKHDRGLRYSYVSNNKQLKGFDVDQTDYLLGIFANGHLKYEHERNKGPEGMPSISDMVEAAIKVLRKNKNGYFLMVEGGNVDMAHHRGWAKRAIDESAALEGAVRLAAAHTDPRDTLLVVTSDHTHTLSINGYPYRGTDIFDIAQTSKYDNVNYTTLSYGTGGPGSMKYSVKTEHNRTVAAREDPGVNTDSFDYQQIAAITLDENSHGGGDVVVYARGPHAHLFHNVHEQHYVYHAISYAAKMGAYSGTSAIYTNISLLAIAAWVAICIMK
Summary
Catalytic Activity
a phosphate monoester + H2O = an alcohol + phosphate
Similarity
Belongs to the alkaline phosphatase family.
Feature
chain Alkaline phosphatase
Uniprot
H9JFV4
A0A2A4K1R0
A0A0F6PQM1
A0A194PCE0
A0A0L7L7G6
A0A3S2LY03
+ More
I6PFJ3 M4M5S9 A0A2H1WHP5 A0A0C9S0U6 A0A3L8DSF8 A0A088AME9 A0A195EYH1 A0A1B6IP91 A0A158NST0 A0A1B6GLC5 A0A1B6ECT8 A0A1B6DR19 E2A827 A0A224XEP7 E2C0T1 T1I0Y8 A0A151X5Q1 A0A1S3CWV3 F4WSI0 A0A151IF46 E0VGW0 A0A310SDV0 A0A2A3EHG5 A0A151I269 A0A232ET19 K7ING2 A0A023F5X3 A0A1L8DK51 N6UBH1 A0A139WDX0 A0A026WZL9 A0A151J5W5 A0A0M9A8U3 A0A0L7RCL8 A0A1B0DA98 A0A2K8JUQ2 A0A1B6DWL1 A0A2R7WWP8 G9FJB7 A0A034VJQ3 A0A0K8U2I9 A0A0A1WTC6 W8BLC1 A0A146KZL8 B4MAX3 A0A0A9X175 A0A3Q0IMU6 B4JKH0 B4L5F8 A0A2L1IQA3 A0A1I8NM94 A0A1S6J0W4 J9K4Y9 A0A0P4VQ65 A0A0M4FAL4 T1PDQ4 B4NCA1 A0A154P1P3 A0A1B0ETD4 B4I5U7 A0A1B0G202 B4Q9X6 Q9VIW9 A8E6R0 B3NM60 B4PAY1 B3MNL4 A0A1W4VF53 A0A0L0BPJ3 A0A0P4W4R7 A0A1W7RB25 A0A2R7VSC7 A0A3R7Q821 A0A2P2I2V2 E9G5V0 A0A087T3L1 A0A0N7ZVC0 K1PG91 A0A164UQ76 A0A0P6DU62 A0A0P5N5D5 A0A0P6GHS2 A0A0N8DZ40 A0A0P5PD28 A0A0P6DU59 A0A0P5S4X1 A0A0N8CA82 A0A0N8EFT7 A0A0P5PQ56 A0A0N8AQD9 A0A0P5NQF6 A0A0N8C2B6
I6PFJ3 M4M5S9 A0A2H1WHP5 A0A0C9S0U6 A0A3L8DSF8 A0A088AME9 A0A195EYH1 A0A1B6IP91 A0A158NST0 A0A1B6GLC5 A0A1B6ECT8 A0A1B6DR19 E2A827 A0A224XEP7 E2C0T1 T1I0Y8 A0A151X5Q1 A0A1S3CWV3 F4WSI0 A0A151IF46 E0VGW0 A0A310SDV0 A0A2A3EHG5 A0A151I269 A0A232ET19 K7ING2 A0A023F5X3 A0A1L8DK51 N6UBH1 A0A139WDX0 A0A026WZL9 A0A151J5W5 A0A0M9A8U3 A0A0L7RCL8 A0A1B0DA98 A0A2K8JUQ2 A0A1B6DWL1 A0A2R7WWP8 G9FJB7 A0A034VJQ3 A0A0K8U2I9 A0A0A1WTC6 W8BLC1 A0A146KZL8 B4MAX3 A0A0A9X175 A0A3Q0IMU6 B4JKH0 B4L5F8 A0A2L1IQA3 A0A1I8NM94 A0A1S6J0W4 J9K4Y9 A0A0P4VQ65 A0A0M4FAL4 T1PDQ4 B4NCA1 A0A154P1P3 A0A1B0ETD4 B4I5U7 A0A1B0G202 B4Q9X6 Q9VIW9 A8E6R0 B3NM60 B4PAY1 B3MNL4 A0A1W4VF53 A0A0L0BPJ3 A0A0P4W4R7 A0A1W7RB25 A0A2R7VSC7 A0A3R7Q821 A0A2P2I2V2 E9G5V0 A0A087T3L1 A0A0N7ZVC0 K1PG91 A0A164UQ76 A0A0P6DU62 A0A0P5N5D5 A0A0P6GHS2 A0A0N8DZ40 A0A0P5PD28 A0A0P6DU59 A0A0P5S4X1 A0A0N8CA82 A0A0N8EFT7 A0A0P5PQ56 A0A0N8AQD9 A0A0P5NQF6 A0A0N8C2B6
EC Number
3.1.3.1
Pubmed
19121390
25662100
26354079
26227816
30249741
21347285
+ More
20798317 21719571 20566863 28648823 20075255 25474469 23537049 18362917 19820115 24508170 22984624 25348373 25830018 24495485 26823975 17994087 18057021 25401762 29415259 27414796 27129103 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26108605 21292972 22992520
20798317 21719571 20566863 28648823 20075255 25474469 23537049 18362917 19820115 24508170 22984624 25348373 25830018 24495485 26823975 17994087 18057021 25401762 29415259 27414796 27129103 25315136 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 26108605 21292972 22992520
EMBL
BABH01001554
NWSH01000253
PCG77999.1
KM244031
AKC42324.1
KQ459606
+ More
KPI90956.1 JTDY01002499 KOB71284.1 RSAL01000124 RVE46598.1 JF979063 AFI81419.1 JQ747497 AGG36454.1 ODYU01008734 SOQ52588.1 GBYB01014231 JAG83998.1 QOIP01000004 RLU23344.1 KQ981923 KYN32957.1 GECU01018965 JAS88741.1 ADTU01025137 GECZ01006515 JAS63254.1 GEDC01001540 JAS35758.1 GEDC01009183 JAS28115.1 GL437468 EFN70422.1 GFTR01006952 JAW09474.1 GL451846 EFN78475.1 ACPB03021812 ACPB03025252 KQ982494 KYQ55733.1 GL888322 EGI62845.1 KQ977808 KYM99548.1 DS235152 EEB12616.1 KQ762882 OAD55347.1 KZ288248 PBC31160.1 KQ976549 KYM80894.1 NNAY01002345 OXU21501.1 GBBI01002119 JAC16593.1 GFDF01007235 JAV06849.1 APGK01041602 APGK01041603 KB740994 KB632255 ENN75992.1 ERL90648.1 KQ971357 KYB26031.1 KK107054 EZA61510.1 KQ979928 KYN18595.1 KQ435711 KOX79580.1 KQ414615 KOC68544.1 AJVK01013241 AJVK01013242 KY031024 ATU82775.1 GEDC01007224 JAS30074.1 KK855543 PTY23055.1 JN135240 AEV66507.1 GAKP01016907 JAC42045.1 GDHF01031340 GDHF01028429 GDHF01028065 GDHF01023895 GDHF01009316 JAI20974.1 JAI23885.1 JAI24249.1 JAI28419.1 JAI42998.1 GBXI01012604 JAD01688.1 GAMC01008737 GAMC01008736 GAMC01008735 JAB97818.1 GDHC01016838 JAQ01791.1 CH940655 EDW66382.1 KRF82644.1 KRF82645.1 GBHO01029882 GBHO01029870 JAG13722.1 JAG13734.1 CH916370 EDW00073.1 CH933811 EDW06417.1 MF741668 AVD96951.1 KU764424 AQS60671.1 ABLF02010432 GDKW01003278 JAI53317.1 CP012528 ALC49837.1 KA646048 AFP60677.1 CH964239 EDW82460.1 KRF99618.1 KQ434796 KZC05781.1 CCAG010014407 CCAG010014408 CH480822 EDW55753.1 CM000361 CM002910 EDX05506.1 KMY91003.1 AE014134 AAF53795.1 ADV37097.1 ADV37098.1 AGB93110.1 AGB93111.1 BT030852 ABV82234.1 CH954179 EDV54660.1 CM000158 EDW90410.1 KRJ99163.1 KRJ99164.1 CH902620 EDV31101.1 JRES01001651 KNC21149.1 GDRN01072395 JAI63554.1 GFAH01000056 JAV48333.1 KK854047 PTY10249.1 QCYY01002370 ROT70910.1 IACF01002718 LAB68362.1 GL732533 EFX85107.1 KK113248 KFM59700.1 GDIP01208977 JAJ14425.1 JH816532 EKC20593.1 LRGB01001579 KZS11573.1 GDIQ01072354 JAN22383.1 GDIQ01146885 GDIQ01146884 GDIQ01145531 JAL04842.1 GDIQ01033013 GDIQ01032023 JAN61724.1 GDIQ01222108 GDIQ01153371 GDIQ01077748 GDIQ01046619 GDIQ01003628 JAN16989.1 GDIQ01130379 GDIQ01130378 GDIQ01061357 JAL21348.1 GDIQ01245354 GDIQ01157767 GDIQ01155573 GDIQ01081759 GDIQ01081758 GDIQ01072353 GDIQ01064619 JAN22384.1 GDIQ01182324 GDIQ01096697 GDIQ01096696 GDIQ01065961 GDIQ01065960 JAL55029.1 GDIQ01099604 GDIQ01099603 GDIQ01063119 JAL52123.1 GDIQ01190561 GDIQ01182323 GDIQ01134377 GDIQ01030972 JAN63765.1 GDIQ01232140 GDIQ01184715 GDIQ01163875 GDIQ01125937 JAL25789.1 GDIQ01253147 GDIQ01245353 GDIQ01169214 JAJ98577.1 GDIQ01144132 GDIQ01141711 JAL10015.1 GDIQ01184717 GDIQ01184716 GDIQ01163874 GDIQ01127531 GDIQ01121731 JAL29995.1
KPI90956.1 JTDY01002499 KOB71284.1 RSAL01000124 RVE46598.1 JF979063 AFI81419.1 JQ747497 AGG36454.1 ODYU01008734 SOQ52588.1 GBYB01014231 JAG83998.1 QOIP01000004 RLU23344.1 KQ981923 KYN32957.1 GECU01018965 JAS88741.1 ADTU01025137 GECZ01006515 JAS63254.1 GEDC01001540 JAS35758.1 GEDC01009183 JAS28115.1 GL437468 EFN70422.1 GFTR01006952 JAW09474.1 GL451846 EFN78475.1 ACPB03021812 ACPB03025252 KQ982494 KYQ55733.1 GL888322 EGI62845.1 KQ977808 KYM99548.1 DS235152 EEB12616.1 KQ762882 OAD55347.1 KZ288248 PBC31160.1 KQ976549 KYM80894.1 NNAY01002345 OXU21501.1 GBBI01002119 JAC16593.1 GFDF01007235 JAV06849.1 APGK01041602 APGK01041603 KB740994 KB632255 ENN75992.1 ERL90648.1 KQ971357 KYB26031.1 KK107054 EZA61510.1 KQ979928 KYN18595.1 KQ435711 KOX79580.1 KQ414615 KOC68544.1 AJVK01013241 AJVK01013242 KY031024 ATU82775.1 GEDC01007224 JAS30074.1 KK855543 PTY23055.1 JN135240 AEV66507.1 GAKP01016907 JAC42045.1 GDHF01031340 GDHF01028429 GDHF01028065 GDHF01023895 GDHF01009316 JAI20974.1 JAI23885.1 JAI24249.1 JAI28419.1 JAI42998.1 GBXI01012604 JAD01688.1 GAMC01008737 GAMC01008736 GAMC01008735 JAB97818.1 GDHC01016838 JAQ01791.1 CH940655 EDW66382.1 KRF82644.1 KRF82645.1 GBHO01029882 GBHO01029870 JAG13722.1 JAG13734.1 CH916370 EDW00073.1 CH933811 EDW06417.1 MF741668 AVD96951.1 KU764424 AQS60671.1 ABLF02010432 GDKW01003278 JAI53317.1 CP012528 ALC49837.1 KA646048 AFP60677.1 CH964239 EDW82460.1 KRF99618.1 KQ434796 KZC05781.1 CCAG010014407 CCAG010014408 CH480822 EDW55753.1 CM000361 CM002910 EDX05506.1 KMY91003.1 AE014134 AAF53795.1 ADV37097.1 ADV37098.1 AGB93110.1 AGB93111.1 BT030852 ABV82234.1 CH954179 EDV54660.1 CM000158 EDW90410.1 KRJ99163.1 KRJ99164.1 CH902620 EDV31101.1 JRES01001651 KNC21149.1 GDRN01072395 JAI63554.1 GFAH01000056 JAV48333.1 KK854047 PTY10249.1 QCYY01002370 ROT70910.1 IACF01002718 LAB68362.1 GL732533 EFX85107.1 KK113248 KFM59700.1 GDIP01208977 JAJ14425.1 JH816532 EKC20593.1 LRGB01001579 KZS11573.1 GDIQ01072354 JAN22383.1 GDIQ01146885 GDIQ01146884 GDIQ01145531 JAL04842.1 GDIQ01033013 GDIQ01032023 JAN61724.1 GDIQ01222108 GDIQ01153371 GDIQ01077748 GDIQ01046619 GDIQ01003628 JAN16989.1 GDIQ01130379 GDIQ01130378 GDIQ01061357 JAL21348.1 GDIQ01245354 GDIQ01157767 GDIQ01155573 GDIQ01081759 GDIQ01081758 GDIQ01072353 GDIQ01064619 JAN22384.1 GDIQ01182324 GDIQ01096697 GDIQ01096696 GDIQ01065961 GDIQ01065960 JAL55029.1 GDIQ01099604 GDIQ01099603 GDIQ01063119 JAL52123.1 GDIQ01190561 GDIQ01182323 GDIQ01134377 GDIQ01030972 JAN63765.1 GDIQ01232140 GDIQ01184715 GDIQ01163875 GDIQ01125937 JAL25789.1 GDIQ01253147 GDIQ01245353 GDIQ01169214 JAJ98577.1 GDIQ01144132 GDIQ01141711 JAL10015.1 GDIQ01184717 GDIQ01184716 GDIQ01163874 GDIQ01127531 GDIQ01121731 JAL29995.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000037510
UP000283053
UP000279307
+ More
UP000005203 UP000078541 UP000005205 UP000000311 UP000008237 UP000015103 UP000075809 UP000079169 UP000007755 UP000078542 UP000009046 UP000242457 UP000078540 UP000215335 UP000002358 UP000019118 UP000030742 UP000007266 UP000053097 UP000078492 UP000053105 UP000053825 UP000092462 UP000008792 UP000001070 UP000009192 UP000095300 UP000007819 UP000092553 UP000095301 UP000007798 UP000076502 UP000092444 UP000001292 UP000000304 UP000000803 UP000008711 UP000002282 UP000007801 UP000192221 UP000037069 UP000283509 UP000000305 UP000054359 UP000005408 UP000076858
UP000005203 UP000078541 UP000005205 UP000000311 UP000008237 UP000015103 UP000075809 UP000079169 UP000007755 UP000078542 UP000009046 UP000242457 UP000078540 UP000215335 UP000002358 UP000019118 UP000030742 UP000007266 UP000053097 UP000078492 UP000053105 UP000053825 UP000092462 UP000008792 UP000001070 UP000009192 UP000095300 UP000007819 UP000092553 UP000095301 UP000007798 UP000076502 UP000092444 UP000001292 UP000000304 UP000000803 UP000008711 UP000002282 UP000007801 UP000192221 UP000037069 UP000283509 UP000000305 UP000054359 UP000005408 UP000076858
Interpro
IPR017850
Alkaline_phosphatase_core_sf
+ More
IPR001952 Alkaline_phosphatase
IPR018299 Alkaline_phosphatase_AS
IPR017452 GPCR_Rhodpsn_7TM
IPR024088 Tyr-tRNA-ligase_bac-type
IPR000276 GPCR_Rhodpsn
IPR002305 aa-tRNA-synth_Ic
IPR002307 Tyr-tRNA-ligase
IPR036986 S4_RNA-bd_sf
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
IPR001952 Alkaline_phosphatase
IPR018299 Alkaline_phosphatase_AS
IPR017452 GPCR_Rhodpsn_7TM
IPR024088 Tyr-tRNA-ligase_bac-type
IPR000276 GPCR_Rhodpsn
IPR002305 aa-tRNA-synth_Ic
IPR002307 Tyr-tRNA-ligase
IPR036986 S4_RNA-bd_sf
IPR014729 Rossmann-like_a/b/a_fold
IPR001412 aa-tRNA-synth_I_CS
SUPFAM
SSF53649
SSF53649
Gene 3D
ProteinModelPortal
H9JFV4
A0A2A4K1R0
A0A0F6PQM1
A0A194PCE0
A0A0L7L7G6
A0A3S2LY03
+ More
I6PFJ3 M4M5S9 A0A2H1WHP5 A0A0C9S0U6 A0A3L8DSF8 A0A088AME9 A0A195EYH1 A0A1B6IP91 A0A158NST0 A0A1B6GLC5 A0A1B6ECT8 A0A1B6DR19 E2A827 A0A224XEP7 E2C0T1 T1I0Y8 A0A151X5Q1 A0A1S3CWV3 F4WSI0 A0A151IF46 E0VGW0 A0A310SDV0 A0A2A3EHG5 A0A151I269 A0A232ET19 K7ING2 A0A023F5X3 A0A1L8DK51 N6UBH1 A0A139WDX0 A0A026WZL9 A0A151J5W5 A0A0M9A8U3 A0A0L7RCL8 A0A1B0DA98 A0A2K8JUQ2 A0A1B6DWL1 A0A2R7WWP8 G9FJB7 A0A034VJQ3 A0A0K8U2I9 A0A0A1WTC6 W8BLC1 A0A146KZL8 B4MAX3 A0A0A9X175 A0A3Q0IMU6 B4JKH0 B4L5F8 A0A2L1IQA3 A0A1I8NM94 A0A1S6J0W4 J9K4Y9 A0A0P4VQ65 A0A0M4FAL4 T1PDQ4 B4NCA1 A0A154P1P3 A0A1B0ETD4 B4I5U7 A0A1B0G202 B4Q9X6 Q9VIW9 A8E6R0 B3NM60 B4PAY1 B3MNL4 A0A1W4VF53 A0A0L0BPJ3 A0A0P4W4R7 A0A1W7RB25 A0A2R7VSC7 A0A3R7Q821 A0A2P2I2V2 E9G5V0 A0A087T3L1 A0A0N7ZVC0 K1PG91 A0A164UQ76 A0A0P6DU62 A0A0P5N5D5 A0A0P6GHS2 A0A0N8DZ40 A0A0P5PD28 A0A0P6DU59 A0A0P5S4X1 A0A0N8CA82 A0A0N8EFT7 A0A0P5PQ56 A0A0N8AQD9 A0A0P5NQF6 A0A0N8C2B6
I6PFJ3 M4M5S9 A0A2H1WHP5 A0A0C9S0U6 A0A3L8DSF8 A0A088AME9 A0A195EYH1 A0A1B6IP91 A0A158NST0 A0A1B6GLC5 A0A1B6ECT8 A0A1B6DR19 E2A827 A0A224XEP7 E2C0T1 T1I0Y8 A0A151X5Q1 A0A1S3CWV3 F4WSI0 A0A151IF46 E0VGW0 A0A310SDV0 A0A2A3EHG5 A0A151I269 A0A232ET19 K7ING2 A0A023F5X3 A0A1L8DK51 N6UBH1 A0A139WDX0 A0A026WZL9 A0A151J5W5 A0A0M9A8U3 A0A0L7RCL8 A0A1B0DA98 A0A2K8JUQ2 A0A1B6DWL1 A0A2R7WWP8 G9FJB7 A0A034VJQ3 A0A0K8U2I9 A0A0A1WTC6 W8BLC1 A0A146KZL8 B4MAX3 A0A0A9X175 A0A3Q0IMU6 B4JKH0 B4L5F8 A0A2L1IQA3 A0A1I8NM94 A0A1S6J0W4 J9K4Y9 A0A0P4VQ65 A0A0M4FAL4 T1PDQ4 B4NCA1 A0A154P1P3 A0A1B0ETD4 B4I5U7 A0A1B0G202 B4Q9X6 Q9VIW9 A8E6R0 B3NM60 B4PAY1 B3MNL4 A0A1W4VF53 A0A0L0BPJ3 A0A0P4W4R7 A0A1W7RB25 A0A2R7VSC7 A0A3R7Q821 A0A2P2I2V2 E9G5V0 A0A087T3L1 A0A0N7ZVC0 K1PG91 A0A164UQ76 A0A0P6DU62 A0A0P5N5D5 A0A0P6GHS2 A0A0N8DZ40 A0A0P5PD28 A0A0P6DU59 A0A0P5S4X1 A0A0N8CA82 A0A0N8EFT7 A0A0P5PQ56 A0A0N8AQD9 A0A0P5NQF6 A0A0N8C2B6
PDB
1K7H
E-value=1.10732e-86,
Score=817
Ontologies
PATHWAY
GO
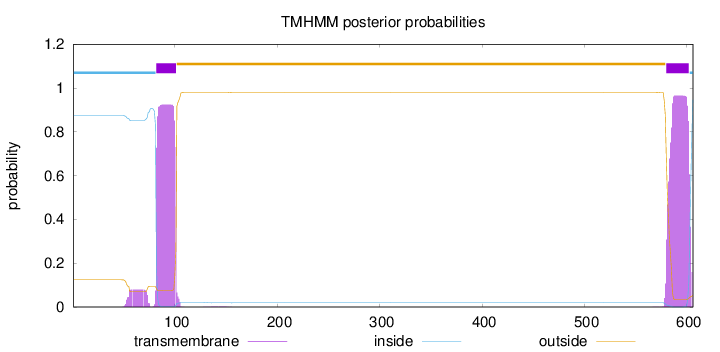
Topology
Length:
606
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
40.87954
Exp number, first 60 AAs:
0.53691
Total prob of N-in:
0.87571
inside
1 - 81
TMhelix
82 - 101
outside
102 - 579
TMhelix
580 - 602
inside
603 - 606
Population Genetic Test Statistics
Pi
212.452048
Theta
158.953738
Tajima's D
1.064755
CLR
0.125705
CSRT
0.67821608919554
Interpretation
Uncertain
