Gene
KWMTBOMO10749
Pre Gene Modal
BGIBMGA008403
Annotation
PREDICTED:_methyltransferase-like_protein_25_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.105
Sequence
CDS
ATGGCTGTAAGTAAGGATTTTAATGACGTATTAGATGACATAGTGCTTGGTGAAGAAAAAGAGAGCAAGTCTAGCTATGATGAAGGGTTCAGAGCAGGCATAGAAGCTGGTAACATAGAGGGTTATCACTTGGGCTATCATCGGGGTGCTGAACTGGGTCGTGAGCTAGGTTTCTACTGGAGCACAGTAAATGAATGCCTTGAACTAAATGAAACATCAGACGCAAAATTATCAGAAAAAATAATAGCCCAGCTGCTCAAAGTTAAAGAACTAATAGATACTTTTCCTCCTAATAATTCTGAAGACCATGACATACTAGGTATGGCCGATGCAGAGTCTCATTCTAAAAGAAAAGATTCAATGAAAACAACAATTCGTAGTCTATGTCACAATATAGATTCCATAATCAACTACCTTGCACCATATTTGCCACTGGTCAACTGTCATATGGTAGAATTCCTAACAAATGACCACTGGAATACATACTTGCCAAATAATTTAAAAGAATATCTCGATCAGCTGGATTTGAATGAGTCACTGGAGCAGTTTTGGAATGCTACTAACAGCTATAAAGGCAATGAACCGACTGAATTGAGAAAATGGATAACGGAGGCCAGAAAACATTATTTGGAAGTCAATAATGACTATTGTTTGTCAGTTGAACAGTTGAATGAAAGGATCAAGTCATGGGGCGGGGAACTGCAGCCAGAGATCAAGGTTAAAGAATTTATGAATAACAAGAAGAGTTATGAAGTACAAACAATGGCACGTGCAGTGTCGTCTGTATACTCGGCGGCATCAGGCACGTGCTGCGTGGAGGCGGGGGGAGGCCGGGGACACTTGCTGACGACCCTAGCGCTCGGGTACCACATACCCAGCCTCACTGTGGATTGCGACCCGAAAACACTAAGTGCTGCTATAAATAGAGTCAAAGTTATACAGAAACAATGGCACGCCATCGCCAAAAGGATACACGACGGAGCCGAAGAGAAACCGCCTCACTTCAATAAAGACCTCCAGAGATTCGCTCAGGCTTTTATAACCAAAGACACTGATCTTGGAGGCGTCGTCAGAGAGATGTTCCCAGAGTGTAAAGACTTGGATACTAGGATTATTCTCACTGGTCTCCATACTTGTGGGAATCTTGGTCCGGACTCGCTTCGTATCTTCACGGCTCGATCGCAGATAGCAGGCCTCCTCACGGTCCCGTGCTGTTACCACCTCCTCACCGAGGACTCAGACGTGGAACTCTTCGATGTTTTCCAGAAGGAGTACGGAACTGGAGAGAGGACGCAGGGTTTCCCGATGTCTGAGTATCTGAGAGGCTACAGGCTGGGTCGCAACGCGCGCATGCTGGCGGCGCAGTCCGTGGACCGCGTGCTGCACCAGCGCCAGCTGCCGGCCCACAGCCTGCTGCACCGGGCCATGCTGCAGGTTGTCATCAAGAAATATTTACCAAACACCACATTAAAGGAAGGCAAATTGAAACGCATCTCGAAATGCGACACCTTCGAACAATACCTCAAGATGGCCGACAAAATTCTTAATTTAAATCTAAATCTGCCCGAAACGTTCTACACAGATACGCAATTGTATTTGGATTGCCAGTGGAAAAAAATGACTTTATTTTATTTAATCAGACTGTGTTTGGCCCAAGTAGTGGAAAGCGTAATATTGTTGGATAGAATATTGTTTTTGTATGAGAACGGCTTCGAGAAAGTTTATTTAGTTAAATTATTTGATCCGGTTTTATCGCCGCGCTGTCACAGTATTGTTGCTTTAAGATAA
Protein
MAVSKDFNDVLDDIVLGEEKESKSSYDEGFRAGIEAGNIEGYHLGYHRGAELGRELGFYWSTVNECLELNETSDAKLSEKIIAQLLKVKELIDTFPPNNSEDHDILGMADAESHSKRKDSMKTTIRSLCHNIDSIINYLAPYLPLVNCHMVEFLTNDHWNTYLPNNLKEYLDQLDLNESLEQFWNATNSYKGNEPTELRKWITEARKHYLEVNNDYCLSVEQLNERIKSWGGELQPEIKVKEFMNNKKSYEVQTMARAVSSVYSAASGTCCVEAGGGRGHLLTTLALGYHIPSLTVDCDPKTLSAAINRVKVIQKQWHAIAKRIHDGAEEKPPHFNKDLQRFAQAFITKDTDLGGVVREMFPECKDLDTRIILTGLHTCGNLGPDSLRIFTARSQIAGLLTVPCCYHLLTEDSDVELFDVFQKEYGTGERTQGFPMSEYLRGYRLGRNARMLAAQSVDRVLHQRQLPAHSLLHRAMLQVVIKKYLPNTTLKEGKLKRISKCDTFEQYLKMADKILNLNLNLPETFYTDTQLYLDCQWKKMTLFYLIRLCLAQVVESVILLDRILFLYENGFEKVYLVKLFDPVLSPRCHSIVALR
Summary
Uniprot
H9JFV6
A0A212F9T8
A0A1E1WNR3
A0A2A4K2Y0
A0A194PD74
A0A139WMX3
+ More
A0A1A9UHH0 A0A1B0G9Z2 A0A336MDL2 A0A1A9WV49 A0A1I8NF28 A0A182WFV0 A0A1B6GWE9 A0A182WUM5 A0A1B6J4W2 A0A182H4B3 A0A0L0C9H5 A0A1B0BSR4 A0A1Q3FJP5 A0A1A9XP82 A0A182K196 B0XAG1 A0A1B6E0U3 A0A1B6MMQ1 B4KTL1 J9JS57 A0A146KZ22 B4LJS8 A0A1A9Z598 A0A0P4VVU0 A0A1I8P9Y8 A0A084WQZ2 A0A0M4EKD1 A0A2H8THJ0 A0A1Y1NKE1 A0A1J1HEU2 Q292C4 B4G9W7 B4HP61 A0A182MX55 B3MC81 A0A182JDJ7 A0A034VQK5 A0A1S3DEY3 A0A1S4EK62 A0A1W4U6E9 A0A2Y9D0H5 E0VYD9 B4NSG1 U4U6R2 B4MY84 A0A182Q4U8 A0A2J7QY44 W5JU37 N6TT61 A0A3B0J5X3 A0A2M4BL67 Q86NX3 A0A2M4BLI2 A0A0A1WHT8 A0A0K8WK41 A0A182YKP2 W8C0B9 A0A2M4BL72 B3NS67 B4P5M7 A0A182KKD2 A0A2M4APW2 B4JW73 A0A182VNE5 A0A182THG6 A0A182HVV0 Q7PFI4 A0A182NA37 A0A0H2UI71 A0A182R2X6 A0A0K8UZV5 A0A2S2PN81 A0A0P6H4U5 A0A1B0DC50 A0A293LLV6 R7UXH0 A0A182PRN2 A0A0P5SP98 A0A0P5BN79 A0A0P5QRE5 A0A0N7ZUF7 A0A0P5VFA0
A0A1A9UHH0 A0A1B0G9Z2 A0A336MDL2 A0A1A9WV49 A0A1I8NF28 A0A182WFV0 A0A1B6GWE9 A0A182WUM5 A0A1B6J4W2 A0A182H4B3 A0A0L0C9H5 A0A1B0BSR4 A0A1Q3FJP5 A0A1A9XP82 A0A182K196 B0XAG1 A0A1B6E0U3 A0A1B6MMQ1 B4KTL1 J9JS57 A0A146KZ22 B4LJS8 A0A1A9Z598 A0A0P4VVU0 A0A1I8P9Y8 A0A084WQZ2 A0A0M4EKD1 A0A2H8THJ0 A0A1Y1NKE1 A0A1J1HEU2 Q292C4 B4G9W7 B4HP61 A0A182MX55 B3MC81 A0A182JDJ7 A0A034VQK5 A0A1S3DEY3 A0A1S4EK62 A0A1W4U6E9 A0A2Y9D0H5 E0VYD9 B4NSG1 U4U6R2 B4MY84 A0A182Q4U8 A0A2J7QY44 W5JU37 N6TT61 A0A3B0J5X3 A0A2M4BL67 Q86NX3 A0A2M4BLI2 A0A0A1WHT8 A0A0K8WK41 A0A182YKP2 W8C0B9 A0A2M4BL72 B3NS67 B4P5M7 A0A182KKD2 A0A2M4APW2 B4JW73 A0A182VNE5 A0A182THG6 A0A182HVV0 Q7PFI4 A0A182NA37 A0A0H2UI71 A0A182R2X6 A0A0K8UZV5 A0A2S2PN81 A0A0P6H4U5 A0A1B0DC50 A0A293LLV6 R7UXH0 A0A182PRN2 A0A0P5SP98 A0A0P5BN79 A0A0P5QRE5 A0A0N7ZUF7 A0A0P5VFA0
Pubmed
19121390
22118469
26354079
18362917
19820115
25315136
+ More
26483478 26108605 17994087 26823975 27129103 24438588 28004739 15632085 25348373 20566863 22936249 23537049 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25244985 24495485 17550304 20966253 12364791 23254933
26483478 26108605 17994087 26823975 27129103 24438588 28004739 15632085 25348373 20566863 22936249 23537049 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25244985 24495485 17550304 20966253 12364791 23254933
EMBL
BABH01001553
AGBW02009561
OWR50502.1
GDQN01011283
GDQN01002466
JAT79771.1
+ More
JAT88588.1 NWSH01000253 PCG78002.1 KQ459606 KPI90958.1 KQ971312 KYB29369.1 CCAG010011403 UFQT01000997 SSX28375.1 GECZ01003032 JAS66737.1 GECU01013512 JAS94194.1 JXUM01109182 KQ565297 KXJ71117.1 JRES01000735 KNC28892.1 JXJN01019826 GFDL01007332 JAV27713.1 DS232581 EDS43658.1 GEDC01005753 JAS31545.1 GEBQ01002794 JAT37183.1 CH933808 EDW09594.1 ABLF02029453 GDHC01016996 JAQ01633.1 CH940648 EDW60587.1 GDKW01000418 JAI56177.1 ATLV01025853 ATLV01025854 ATLV01025855 KE525401 KFB52636.1 CP012524 ALC41830.1 GFXV01001778 MBW13583.1 GEZM01002890 JAV97085.1 CVRI01000001 CRK86355.1 CM000071 EAL24938.2 CH479181 EDW31719.1 CH480816 EDW47510.1 AXCM01000462 CH902619 EDV36181.1 GAKP01013366 JAC45586.1 DS235845 EEB18395.1 CH982209 CM002911 EDX15539.1 KMY93116.1 KB632173 ERL89579.1 CH963894 EDW77073.1 AXCN02002379 NEVH01009368 PNF33505.1 ADMH02000135 ETN67646.1 APGK01019954 KB740150 ENN81253.1 OUUW01000001 SPP75112.1 GGFJ01004679 MBW53820.1 AE013599 BT003595 AAF58558.4 AAO39598.1 GGFJ01004680 MBW53821.1 GBXI01015870 JAC98421.1 GDHF01000798 JAI51516.1 GAMC01004102 JAC02454.1 GGFJ01004676 MBW53817.1 CH954179 EDV56369.1 CM000158 EDW90824.1 GGFK01009488 MBW42809.1 CH916375 EDV98211.1 APCN01001422 AAAB01008823 EAA45304.4 ACPB03006429 GDHF01020112 JAI32202.1 GGMR01018246 MBY30865.1 GDIQ01024458 JAN70279.1 AJVK01005022 AJVK01005023 GFWV01004108 MAA28838.1 AMQN01005883 KB297068 ELU10992.1 GDIP01137236 JAL66478.1 GDIP01182639 JAJ40763.1 GDIQ01110584 JAL41142.1 GDIP01211481 GDIP01133634 JAJ11921.1 GDIP01102547 JAM01168.1
JAT88588.1 NWSH01000253 PCG78002.1 KQ459606 KPI90958.1 KQ971312 KYB29369.1 CCAG010011403 UFQT01000997 SSX28375.1 GECZ01003032 JAS66737.1 GECU01013512 JAS94194.1 JXUM01109182 KQ565297 KXJ71117.1 JRES01000735 KNC28892.1 JXJN01019826 GFDL01007332 JAV27713.1 DS232581 EDS43658.1 GEDC01005753 JAS31545.1 GEBQ01002794 JAT37183.1 CH933808 EDW09594.1 ABLF02029453 GDHC01016996 JAQ01633.1 CH940648 EDW60587.1 GDKW01000418 JAI56177.1 ATLV01025853 ATLV01025854 ATLV01025855 KE525401 KFB52636.1 CP012524 ALC41830.1 GFXV01001778 MBW13583.1 GEZM01002890 JAV97085.1 CVRI01000001 CRK86355.1 CM000071 EAL24938.2 CH479181 EDW31719.1 CH480816 EDW47510.1 AXCM01000462 CH902619 EDV36181.1 GAKP01013366 JAC45586.1 DS235845 EEB18395.1 CH982209 CM002911 EDX15539.1 KMY93116.1 KB632173 ERL89579.1 CH963894 EDW77073.1 AXCN02002379 NEVH01009368 PNF33505.1 ADMH02000135 ETN67646.1 APGK01019954 KB740150 ENN81253.1 OUUW01000001 SPP75112.1 GGFJ01004679 MBW53820.1 AE013599 BT003595 AAF58558.4 AAO39598.1 GGFJ01004680 MBW53821.1 GBXI01015870 JAC98421.1 GDHF01000798 JAI51516.1 GAMC01004102 JAC02454.1 GGFJ01004676 MBW53817.1 CH954179 EDV56369.1 CM000158 EDW90824.1 GGFK01009488 MBW42809.1 CH916375 EDV98211.1 APCN01001422 AAAB01008823 EAA45304.4 ACPB03006429 GDHF01020112 JAI32202.1 GGMR01018246 MBY30865.1 GDIQ01024458 JAN70279.1 AJVK01005022 AJVK01005023 GFWV01004108 MAA28838.1 AMQN01005883 KB297068 ELU10992.1 GDIP01137236 JAL66478.1 GDIP01182639 JAJ40763.1 GDIQ01110584 JAL41142.1 GDIP01211481 GDIP01133634 JAJ11921.1 GDIP01102547 JAM01168.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000007266
UP000078200
+ More
UP000092444 UP000091820 UP000095301 UP000075920 UP000076407 UP000069940 UP000249989 UP000037069 UP000092460 UP000092443 UP000075881 UP000002320 UP000009192 UP000007819 UP000008792 UP000092445 UP000095300 UP000030765 UP000092553 UP000183832 UP000001819 UP000008744 UP000001292 UP000075883 UP000007801 UP000075880 UP000079169 UP000192221 UP000069272 UP000009046 UP000000304 UP000030742 UP000007798 UP000075886 UP000235965 UP000000673 UP000019118 UP000268350 UP000000803 UP000076408 UP000008711 UP000002282 UP000075882 UP000001070 UP000075903 UP000075902 UP000075840 UP000007062 UP000075884 UP000015103 UP000075900 UP000092462 UP000014760 UP000075885
UP000092444 UP000091820 UP000095301 UP000075920 UP000076407 UP000069940 UP000249989 UP000037069 UP000092460 UP000092443 UP000075881 UP000002320 UP000009192 UP000007819 UP000008792 UP000092445 UP000095300 UP000030765 UP000092553 UP000183832 UP000001819 UP000008744 UP000001292 UP000075883 UP000007801 UP000075880 UP000079169 UP000192221 UP000069272 UP000009046 UP000000304 UP000030742 UP000007798 UP000075886 UP000235965 UP000000673 UP000019118 UP000268350 UP000000803 UP000076408 UP000008711 UP000002282 UP000075882 UP000001070 UP000075903 UP000075902 UP000075840 UP000007062 UP000075884 UP000015103 UP000075900 UP000092462 UP000014760 UP000075885
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9JFV6
A0A212F9T8
A0A1E1WNR3
A0A2A4K2Y0
A0A194PD74
A0A139WMX3
+ More
A0A1A9UHH0 A0A1B0G9Z2 A0A336MDL2 A0A1A9WV49 A0A1I8NF28 A0A182WFV0 A0A1B6GWE9 A0A182WUM5 A0A1B6J4W2 A0A182H4B3 A0A0L0C9H5 A0A1B0BSR4 A0A1Q3FJP5 A0A1A9XP82 A0A182K196 B0XAG1 A0A1B6E0U3 A0A1B6MMQ1 B4KTL1 J9JS57 A0A146KZ22 B4LJS8 A0A1A9Z598 A0A0P4VVU0 A0A1I8P9Y8 A0A084WQZ2 A0A0M4EKD1 A0A2H8THJ0 A0A1Y1NKE1 A0A1J1HEU2 Q292C4 B4G9W7 B4HP61 A0A182MX55 B3MC81 A0A182JDJ7 A0A034VQK5 A0A1S3DEY3 A0A1S4EK62 A0A1W4U6E9 A0A2Y9D0H5 E0VYD9 B4NSG1 U4U6R2 B4MY84 A0A182Q4U8 A0A2J7QY44 W5JU37 N6TT61 A0A3B0J5X3 A0A2M4BL67 Q86NX3 A0A2M4BLI2 A0A0A1WHT8 A0A0K8WK41 A0A182YKP2 W8C0B9 A0A2M4BL72 B3NS67 B4P5M7 A0A182KKD2 A0A2M4APW2 B4JW73 A0A182VNE5 A0A182THG6 A0A182HVV0 Q7PFI4 A0A182NA37 A0A0H2UI71 A0A182R2X6 A0A0K8UZV5 A0A2S2PN81 A0A0P6H4U5 A0A1B0DC50 A0A293LLV6 R7UXH0 A0A182PRN2 A0A0P5SP98 A0A0P5BN79 A0A0P5QRE5 A0A0N7ZUF7 A0A0P5VFA0
A0A1A9UHH0 A0A1B0G9Z2 A0A336MDL2 A0A1A9WV49 A0A1I8NF28 A0A182WFV0 A0A1B6GWE9 A0A182WUM5 A0A1B6J4W2 A0A182H4B3 A0A0L0C9H5 A0A1B0BSR4 A0A1Q3FJP5 A0A1A9XP82 A0A182K196 B0XAG1 A0A1B6E0U3 A0A1B6MMQ1 B4KTL1 J9JS57 A0A146KZ22 B4LJS8 A0A1A9Z598 A0A0P4VVU0 A0A1I8P9Y8 A0A084WQZ2 A0A0M4EKD1 A0A2H8THJ0 A0A1Y1NKE1 A0A1J1HEU2 Q292C4 B4G9W7 B4HP61 A0A182MX55 B3MC81 A0A182JDJ7 A0A034VQK5 A0A1S3DEY3 A0A1S4EK62 A0A1W4U6E9 A0A2Y9D0H5 E0VYD9 B4NSG1 U4U6R2 B4MY84 A0A182Q4U8 A0A2J7QY44 W5JU37 N6TT61 A0A3B0J5X3 A0A2M4BL67 Q86NX3 A0A2M4BLI2 A0A0A1WHT8 A0A0K8WK41 A0A182YKP2 W8C0B9 A0A2M4BL72 B3NS67 B4P5M7 A0A182KKD2 A0A2M4APW2 B4JW73 A0A182VNE5 A0A182THG6 A0A182HVV0 Q7PFI4 A0A182NA37 A0A0H2UI71 A0A182R2X6 A0A0K8UZV5 A0A2S2PN81 A0A0P6H4U5 A0A1B0DC50 A0A293LLV6 R7UXH0 A0A182PRN2 A0A0P5SP98 A0A0P5BN79 A0A0P5QRE5 A0A0N7ZUF7 A0A0P5VFA0
Ontologies
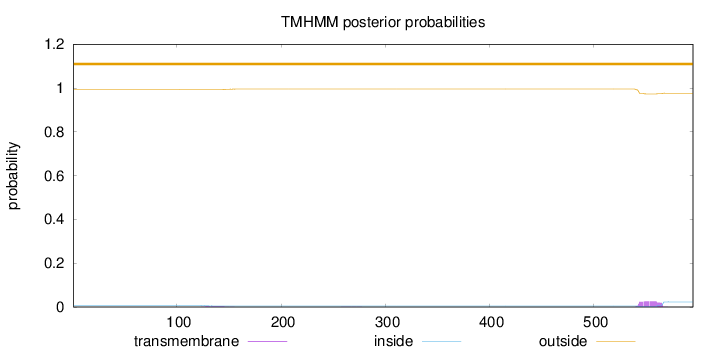
Topology
Length:
595
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.635749999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00760
outside
1 - 595
Population Genetic Test Statistics
Pi
168.072791
Theta
160.262456
Tajima's D
0.630938
CLR
0.089576
CSRT
0.555372231388431
Interpretation
Uncertain
