Gene
KWMTBOMO10742
Annotation
Zinc_finger_MYM-type_protein_1?_partial_[Stegodyphus_mimosarum]
Location in the cell
Cytoplasmic Reliability : 1.146 Nuclear Reliability : 1.52
Sequence
CDS
ATGAAACGAGTAAAAGAAAGCGGTGCGTCATTTAGAAAGAAAATAAAGAAAAGGGAGGAAATACAGATTAAAAACACTGGAGCACTACTCAAATACATCTCTCATAAGCCAACTTCTAGTTCTTTTAGTATTGTTGCCTCAAATACGGGTGAAGAAGCAGCGCTCGAAGGCGAAAAAACATTATCCAATGTTGTTCAAGAGGAAAGACCTGCTTCACACAATGGCGAAGAAGTAGAAAGTTTATCAAGTCCTGATGAAGAATCTGCTGCACAAATTATTCTGTCTGTTTCTATGAAAGATATTGGATTATGGCCTAACAAAATTGACGACAATACAAGAGTATTTCTGGTACGTCAAGGTCCCTTTTTAATTCAAAATTTAGACGCCGACTTCAGCGAAGGAATAAAACGTTCTAATAATTCAAGCAAAGCCAAAGGTGAAACAAGAAAACTGAGTCGGGACTGGTTTTTTCAAATTTTGCCTAATGGCGAAAAGCTCTTAAGATCGTGGATGGTATATTCACCATCGAAGAAAAGTATTTATTGTTTCTGCTGCAAACTGTTTGATAATCTTTGTCAAGCAGGTTTTAGTACGGAAAATGGTTTTAACAAATGGTGGAAACTTAATCCTAAAATTAGTCAACATGAGGTTAGTCCGCTGCACATTCGATCATATTACAAATGGAAGGAATTAGAAATAAGACTTGGAAGTGGAGCAACTATTGACAAACTACAACAACAGAATATTCATAAAGAAATAAAAAAATGGAAGGAAATTCTAACAAGAATACTTGATATAATTAGATTCTTAGCAAAACAAAATTTGGCATTTCGAGGACATCGTGAAAGTGTTCATCAGGAAAACATCGATTTACTGGAAAATAGAGGAAATTTTCTCGAGGTTGTTGACTTATTGGCTAAGTACGACCCAGTATTACGAGAACATGTAGTAAAAATTAAGATGGGTAATAAATATAGTCTCTCTTATTTGTCGCCGAAGATTCAGAACGAATTCATAGAGCTATTGGGAGGCGCAGTCAGAAAAACAATTATGGATCAAATAAAGCAGGCTAAATACTTTTGTATTATATTTGATAGCACTCCTGACATTTCACACAAAGATCAAACCAGTCAAATTATTAGATATGTAGTTATTGAAGGTAGCGAAGTTAAAGTTGTAGAATTGTTTATTGATTTTGTAGAAACTAAAGGAAAAACTGCTGTTGAAATCACAGATATGATTTTAACGAAATTAGAAAATGACGGTTTAGATATAAAAAATTGCAGAGGACAGGCTTACGATAATGCTGCCGTTATGGCAGGAAAGCACAGCGGTGTTCAAACTCGCATTAAAGAAATAAATCCAAATGCAGAATTTGTAGCATGTACAAATCATTCCTTAAACCTGGCATGTTTGCACGCAGCTTCTACAGCTATCAATTCCATCACATTTTTTGGAACTATCGAACAGTTGTTTTCGTTGTTTCCTTCTTCCACTCATAGATGGAATGTTTTACTTTCGGTCACTGGTCAAGGTGTTAAACGTAGTGTAGAAACTCGCTGGAGCGCTAGAGCTGCCGCGTGTCGCTAG
Protein
MKRVKESGASFRKKIKKREEIQIKNTGALLKYISHKPTSSSFSIVASNTGEEAALEGEKTLSNVVQEERPASHNGEEVESLSSPDEESAAQIILSVSMKDIGLWPNKIDDNTRVFLVRQGPFLIQNLDADFSEGIKRSNNSSKAKGETRKLSRDWFFQILPNGEKLLRSWMVYSPSKKSIYCFCCKLFDNLCQAGFSTENGFNKWWKLNPKISQHEVSPLHIRSYYKWKELEIRLGSGATIDKLQQQNIHKEIKKWKEILTRILDIIRFLAKQNLAFRGHRESVHQENIDLLENRGNFLEVVDLLAKYDPVLREHVVKIKMGNKYSLSYLSPKIQNEFIELLGGAVRKTIMDQIKQAKYFCIIFDSTPDISHKDQTSQIIRYVVIEGSEVKVVELFIDFVETKGKTAVEITDMILTKLENDGLDIKNCRGQAYDNAAVMAGKHSGVQTRIKEINPNAEFVACTNHSLNLACLHAASTAINSITFFGTIEQLFSLFPSSTHRWNVLLSVTGQGVKRSVETRWSARAAACR
Summary
Uniprot
A0A087TDI3
A0A087UAK3
A0A2H1V0B8
A0A0L8HCR5
A0A1B6DAI5
A0A151IAV6
+ More
A0A2S2QTP2 A0A2H8TX80 J9M5K2 A0A3S2LR27 A0A0F8B336 A0A151JR63 A0A0F8ARZ7 A0A151JQK9 A0A2S2PRM7 A0A0J7K9X2 I3KVY0 A0A3P8P7S0 A0A087TIQ9 H3ATL5 A0A3P8P3G9 A6BLM9 A0A3P8NJ49 H3ANM7 A0A3P8P7T1 A0A3P9BQ91 A0A3P8PCN0 A0A3P8R8S5 A0A3P9CFP0 A0A2S2NPI7 A0A087TT59 A0A087TDZ4 A0A087TNS4 T2MBP3 A0A2N9HTT2 A0A3L8DD89 A0A2N9IUV3 A0A1D6M0J2 A0A2U1LR28 A0A2N9J138 Q0E2I3 A0A2N9I0X3 A0A1D6MAK2 A0A2N9IRP9 A0A2N9IXV5 A0A2N9EIY1 A0A2N9F6X1 A0A2N9IJF9 A0A2N9J2P4 A0A226D874 A0A1Y1MZW7 A0A2N9HPT9 X1XM94 A0A251T6Q9 A0A3B1J5G5 A0A3S2PND3 A0A3B3C8M6 A0A2N9IH69 A0A2N9FLN4 A0A0P6D2Y9 A0A2N9FYI5 A0A0P6EBA1 A0A2S2QLG5 H3AK32 A0A1D6KSA8 A0A2N9HCB6 A0A3P9LLA6 A0A1A8U928 J9K1R9 A0A1D6ML06 A0A1D6HCG9 A0A2U1M0J0 A0A2N9HC04 D0UZH7 A0A251THY1 A2X9N5 A0A3P9BDZ2 A0A0D3AWL3 A0A226DE12 A0A3L6F912 A0A2K2DE26 A0A3L6FM27 A0A226DET0 A0A2T8KV52 A0A1L8GBW2 T1MRQ8 A0A0P4XYH4
A0A2S2QTP2 A0A2H8TX80 J9M5K2 A0A3S2LR27 A0A0F8B336 A0A151JR63 A0A0F8ARZ7 A0A151JQK9 A0A2S2PRM7 A0A0J7K9X2 I3KVY0 A0A3P8P7S0 A0A087TIQ9 H3ATL5 A0A3P8P3G9 A6BLM9 A0A3P8NJ49 H3ANM7 A0A3P8P7T1 A0A3P9BQ91 A0A3P8PCN0 A0A3P8R8S5 A0A3P9CFP0 A0A2S2NPI7 A0A087TT59 A0A087TDZ4 A0A087TNS4 T2MBP3 A0A2N9HTT2 A0A3L8DD89 A0A2N9IUV3 A0A1D6M0J2 A0A2U1LR28 A0A2N9J138 Q0E2I3 A0A2N9I0X3 A0A1D6MAK2 A0A2N9IRP9 A0A2N9IXV5 A0A2N9EIY1 A0A2N9F6X1 A0A2N9IJF9 A0A2N9J2P4 A0A226D874 A0A1Y1MZW7 A0A2N9HPT9 X1XM94 A0A251T6Q9 A0A3B1J5G5 A0A3S2PND3 A0A3B3C8M6 A0A2N9IH69 A0A2N9FLN4 A0A0P6D2Y9 A0A2N9FYI5 A0A0P6EBA1 A0A2S2QLG5 H3AK32 A0A1D6KSA8 A0A2N9HCB6 A0A3P9LLA6 A0A1A8U928 J9K1R9 A0A1D6ML06 A0A1D6HCG9 A0A2U1M0J0 A0A2N9HC04 D0UZH7 A0A251THY1 A2X9N5 A0A3P9BDZ2 A0A0D3AWL3 A0A226DE12 A0A3L6F912 A0A2K2DE26 A0A3L6FM27 A0A226DET0 A0A2T8KV52 A0A1L8GBW2 T1MRQ8 A0A0P4XYH4
Pubmed
EMBL
KK114731
KFM63172.1
KK119011
KFM74392.1
ODYU01000103
SOQ34303.1
+ More
KQ418495 KOF87093.1 GEDC01014594 JAS22704.1 KQ978156 KYM96678.1 GGMS01011279 MBY80482.1 GFXV01006596 MBW18401.1 ABLF02006249 ABLF02066000 RSAL01000315 RVE42619.1 KQ041811 KKF22417.1 KQ978627 KYN29561.1 KQ040999 KKF32005.1 KQ978697 KYN29158.1 GGMR01019454 MBY32073.1 LBMM01011085 KMQ87036.1 AERX01072813 KK115395 KFM64998.1 AFYH01135549 AB264112 AB288091 BAF64515.1 BAF64516.1 AFYH01177734 GGMR01006047 MBY18666.1 KK116626 KFM68298.1 KK114788 KFM63333.1 KK116090 KFM66763.1 HAAD01003421 CDG69653.1 OIVN01004148 SPD15752.1 QOIP01000009 RLU18344.1 OIVN01006240 SPD28662.1 CM000782 AQK84823.1 PKPP01008157 PWA51452.1 OIVN01006304 SPD30149.1 AP008208 BAF08305.2 OIVN01004480 SPD17674.1 AQK87807.1 OIVN01006223 SPD28157.1 OIVN01006283 SPD29722.1 OIVN01000125 SPC74763.1 OIVN01000591 SPC82609.1 OIVN01005857 SPD24260.1 OIVN01006325 SPD30591.1 LNIX01000031 OXA40937.1 GEZM01016663 GEZM01016662 JAV91212.1 OIVN01003814 SPD13763.1 ABLF02014880 CM007900 OTG06594.1 CM012442 RVE71391.1 OIVN01005696 SPD23655.1 OIVN01001246 SPC91696.1 GDIQ01083731 JAN11006.1 OIVN01001273 SPC92021.1 GDIQ01065287 JAN29450.1 GGMS01009402 MBY78605.1 AFYH01168712 CM007647 ONM05560.1 OIVN01003170 SPD09271.1 HAEJ01004347 SBS44804.1 ABLF02038233 ABLF02038237 CM007649 ONM29894.1 CM000781 AQK72395.1 PKPP01006964 PWA54786.1 SPD09273.1 GQ984160 ACX94087.1 CM007899 OTG10363.1 CM000127 EAY87545.1 LNIX01000023 OXA43094.1 NCVQ01000005 PWZ29694.1 CM000881 PNT72528.1 NCVQ01000004 PWZ34246.1 OXA43091.1 CM008046 PVH66044.1 CM004474 OCT81265.1 GDIP01236011 JAI87390.1
KQ418495 KOF87093.1 GEDC01014594 JAS22704.1 KQ978156 KYM96678.1 GGMS01011279 MBY80482.1 GFXV01006596 MBW18401.1 ABLF02006249 ABLF02066000 RSAL01000315 RVE42619.1 KQ041811 KKF22417.1 KQ978627 KYN29561.1 KQ040999 KKF32005.1 KQ978697 KYN29158.1 GGMR01019454 MBY32073.1 LBMM01011085 KMQ87036.1 AERX01072813 KK115395 KFM64998.1 AFYH01135549 AB264112 AB288091 BAF64515.1 BAF64516.1 AFYH01177734 GGMR01006047 MBY18666.1 KK116626 KFM68298.1 KK114788 KFM63333.1 KK116090 KFM66763.1 HAAD01003421 CDG69653.1 OIVN01004148 SPD15752.1 QOIP01000009 RLU18344.1 OIVN01006240 SPD28662.1 CM000782 AQK84823.1 PKPP01008157 PWA51452.1 OIVN01006304 SPD30149.1 AP008208 BAF08305.2 OIVN01004480 SPD17674.1 AQK87807.1 OIVN01006223 SPD28157.1 OIVN01006283 SPD29722.1 OIVN01000125 SPC74763.1 OIVN01000591 SPC82609.1 OIVN01005857 SPD24260.1 OIVN01006325 SPD30591.1 LNIX01000031 OXA40937.1 GEZM01016663 GEZM01016662 JAV91212.1 OIVN01003814 SPD13763.1 ABLF02014880 CM007900 OTG06594.1 CM012442 RVE71391.1 OIVN01005696 SPD23655.1 OIVN01001246 SPC91696.1 GDIQ01083731 JAN11006.1 OIVN01001273 SPC92021.1 GDIQ01065287 JAN29450.1 GGMS01009402 MBY78605.1 AFYH01168712 CM007647 ONM05560.1 OIVN01003170 SPD09271.1 HAEJ01004347 SBS44804.1 ABLF02038233 ABLF02038237 CM007649 ONM29894.1 CM000781 AQK72395.1 PKPP01006964 PWA54786.1 SPD09273.1 GQ984160 ACX94087.1 CM007899 OTG10363.1 CM000127 EAY87545.1 LNIX01000023 OXA43094.1 NCVQ01000005 PWZ29694.1 CM000881 PNT72528.1 NCVQ01000004 PWZ34246.1 OXA43091.1 CM008046 PVH66044.1 CM004474 OCT81265.1 GDIP01236011 JAI87390.1
Proteomes
UP000054359
UP000053454
UP000078542
UP000007819
UP000283053
UP000078492
+ More
UP000036403 UP000005207 UP000265100 UP000008672 UP000001038 UP000265160 UP000279307 UP000007305 UP000245207 UP000198287 UP000215914 UP000018467 UP000261560 UP000265180 UP000007015 UP000032141 UP000251960 UP000008810 UP000243499 UP000186698 UP000015106
UP000036403 UP000005207 UP000265100 UP000008672 UP000001038 UP000265160 UP000279307 UP000007305 UP000245207 UP000198287 UP000215914 UP000018467 UP000261560 UP000265180 UP000007015 UP000032141 UP000251960 UP000008810 UP000243499 UP000186698 UP000015106
Interpro
Gene 3D
ProteinModelPortal
A0A087TDI3
A0A087UAK3
A0A2H1V0B8
A0A0L8HCR5
A0A1B6DAI5
A0A151IAV6
+ More
A0A2S2QTP2 A0A2H8TX80 J9M5K2 A0A3S2LR27 A0A0F8B336 A0A151JR63 A0A0F8ARZ7 A0A151JQK9 A0A2S2PRM7 A0A0J7K9X2 I3KVY0 A0A3P8P7S0 A0A087TIQ9 H3ATL5 A0A3P8P3G9 A6BLM9 A0A3P8NJ49 H3ANM7 A0A3P8P7T1 A0A3P9BQ91 A0A3P8PCN0 A0A3P8R8S5 A0A3P9CFP0 A0A2S2NPI7 A0A087TT59 A0A087TDZ4 A0A087TNS4 T2MBP3 A0A2N9HTT2 A0A3L8DD89 A0A2N9IUV3 A0A1D6M0J2 A0A2U1LR28 A0A2N9J138 Q0E2I3 A0A2N9I0X3 A0A1D6MAK2 A0A2N9IRP9 A0A2N9IXV5 A0A2N9EIY1 A0A2N9F6X1 A0A2N9IJF9 A0A2N9J2P4 A0A226D874 A0A1Y1MZW7 A0A2N9HPT9 X1XM94 A0A251T6Q9 A0A3B1J5G5 A0A3S2PND3 A0A3B3C8M6 A0A2N9IH69 A0A2N9FLN4 A0A0P6D2Y9 A0A2N9FYI5 A0A0P6EBA1 A0A2S2QLG5 H3AK32 A0A1D6KSA8 A0A2N9HCB6 A0A3P9LLA6 A0A1A8U928 J9K1R9 A0A1D6ML06 A0A1D6HCG9 A0A2U1M0J0 A0A2N9HC04 D0UZH7 A0A251THY1 A2X9N5 A0A3P9BDZ2 A0A0D3AWL3 A0A226DE12 A0A3L6F912 A0A2K2DE26 A0A3L6FM27 A0A226DET0 A0A2T8KV52 A0A1L8GBW2 T1MRQ8 A0A0P4XYH4
A0A2S2QTP2 A0A2H8TX80 J9M5K2 A0A3S2LR27 A0A0F8B336 A0A151JR63 A0A0F8ARZ7 A0A151JQK9 A0A2S2PRM7 A0A0J7K9X2 I3KVY0 A0A3P8P7S0 A0A087TIQ9 H3ATL5 A0A3P8P3G9 A6BLM9 A0A3P8NJ49 H3ANM7 A0A3P8P7T1 A0A3P9BQ91 A0A3P8PCN0 A0A3P8R8S5 A0A3P9CFP0 A0A2S2NPI7 A0A087TT59 A0A087TDZ4 A0A087TNS4 T2MBP3 A0A2N9HTT2 A0A3L8DD89 A0A2N9IUV3 A0A1D6M0J2 A0A2U1LR28 A0A2N9J138 Q0E2I3 A0A2N9I0X3 A0A1D6MAK2 A0A2N9IRP9 A0A2N9IXV5 A0A2N9EIY1 A0A2N9F6X1 A0A2N9IJF9 A0A2N9J2P4 A0A226D874 A0A1Y1MZW7 A0A2N9HPT9 X1XM94 A0A251T6Q9 A0A3B1J5G5 A0A3S2PND3 A0A3B3C8M6 A0A2N9IH69 A0A2N9FLN4 A0A0P6D2Y9 A0A2N9FYI5 A0A0P6EBA1 A0A2S2QLG5 H3AK32 A0A1D6KSA8 A0A2N9HCB6 A0A3P9LLA6 A0A1A8U928 J9K1R9 A0A1D6ML06 A0A1D6HCG9 A0A2U1M0J0 A0A2N9HC04 D0UZH7 A0A251THY1 A2X9N5 A0A3P9BDZ2 A0A0D3AWL3 A0A226DE12 A0A3L6F912 A0A2K2DE26 A0A3L6FM27 A0A226DET0 A0A2T8KV52 A0A1L8GBW2 T1MRQ8 A0A0P4XYH4
Ontologies
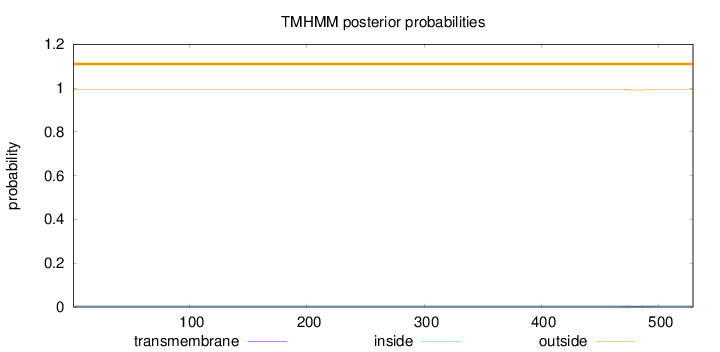
Topology
Length:
529
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.08415
Exp number, first 60 AAs:
0.00074
Total prob of N-in:
0.00667
outside
1 - 529
Population Genetic Test Statistics
Pi
475.912786
Theta
200.649233
Tajima's D
4.392519
CLR
2.39805
CSRT
0.999650017499125
Interpretation
Possibly Balancing Selection
