Gene
KWMTBOMO10741
Annotation
PREDICTED:_putative_odorant_receptor_85d_[Bombyx_mori]
Full name
Odorant receptor
Location in the cell
Mitochondrial Reliability : 1.016 PlasmaMembrane Reliability : 1.324
Sequence
CDS
ATGGCCGGTAGACAGATAAAGAGCTCTAACATTTCTGACGCCGCTTACAACAGCCTCTGGTATGAAGGCGACCTCAAACTTAGAAAACTAATGTTGTTTATCATCACTCGATCACAAAATCCGTGCTATTTATCTGCATTGGGATTTTCAAATATGACTCTACGATCATTTTCAAAGATTATGAGCACAGCATGGACGTATCTGTCGCTTCTTATTCAAGTTTATGAAGGAAATTAA
Protein
MAGRQIKSSNISDAAYNSLWYEGDLKLRKLMLFIITRSQNPCYLSALGFSNMTLRSFSKIMSTAWTYLSLLIQVYEGN
Summary
Similarity
Belongs to the insect chemoreceptor superfamily. Heteromeric odorant receptor channel (TC 1.A.69) family.
Uniprot
A0A2A4K1Q7
A0A2A4K2M3
A0A075T7Y9
A0A2L1TG45
A0A2A4J2I8
A0A386H9E1
+ More
A0A0K8TUP9 A0A223HD29 A0A223HD35 A0A1Y9TJS3 A0A1B3B766 A0A0V0J0N8 C4B7V6 A7E3G8 A0A097ITZ3 A0A0K8TU41 A0A097IYK3 A0A1B2G2P4 A0A0B5CSU1 A0A345BEY2 A0A0B5GQ56 A0A223HDH0 A0A1V1WC59 A0A146JVN1 A0A2L1TG60 A0A2K8GKT2 A0A0V0J2Q1 A0A2K8GKR7 A0A1B3P5N0 A0A0N1IA66 A0A223HD84 A0A2K8GL49 A0A3G6V789 A0A223HDF3 A0A2H1VMQ9 A0A0P1IW06 A0A0P1IVT0 A0A2H1VAZ7 A0A0P1J430 A0A1V1WBS9 A0A0S1TL39 A0A2W1BUH6 A0A1S5XXM4 A0A0U2QG96 A0A2K8GL39 A0A076E991 Q8MMH9 A0A076E9C7 A0A0E4B427 A0A2A4K7V7 A0A146JXZ9 A0A146JYW4 C4B7V5 A0A0U2SQV8 A0A2H1V1Z3 A0A0E3VLQ3 A0A0P1IW02 A0A0E3VLQ7 A0A0E4B3V9 A0A2L1TG41 A0A2L1TG32 A0A0N7MCA7 U5NFR2 A0A386H8Y9 A0A1B3P5U6 A0A1S5XXP8 A0A1E1WPI5 A0A0B4ZWC0 Q6A1K2 A0A2L1TG21 A0A1B3B722 A0A3S2NXV2 R4JKH1 B5A7F1 A0A345BEY3 A0A3S2P9E5 A0A172QGF4 A0A075T622 R4JLY8 A0A1B2G2N4 A0A172QGF5 A0A2K8GKR8 R4JUT8 R4JSF7 A0A2W1BDT5 A0A2A4K9L1 A0A2H1V3Y7 A0A1B3P5M4 A0A1V1WBW6 A0A1B0GLR5 A0A0S1TPW8 A0A1V1WC75 A0A0S1TKX5 A0A1B3B742 A0A2K8GKS1
A0A0K8TUP9 A0A223HD29 A0A223HD35 A0A1Y9TJS3 A0A1B3B766 A0A0V0J0N8 C4B7V6 A7E3G8 A0A097ITZ3 A0A0K8TU41 A0A097IYK3 A0A1B2G2P4 A0A0B5CSU1 A0A345BEY2 A0A0B5GQ56 A0A223HDH0 A0A1V1WC59 A0A146JVN1 A0A2L1TG60 A0A2K8GKT2 A0A0V0J2Q1 A0A2K8GKR7 A0A1B3P5N0 A0A0N1IA66 A0A223HD84 A0A2K8GL49 A0A3G6V789 A0A223HDF3 A0A2H1VMQ9 A0A0P1IW06 A0A0P1IVT0 A0A2H1VAZ7 A0A0P1J430 A0A1V1WBS9 A0A0S1TL39 A0A2W1BUH6 A0A1S5XXM4 A0A0U2QG96 A0A2K8GL39 A0A076E991 Q8MMH9 A0A076E9C7 A0A0E4B427 A0A2A4K7V7 A0A146JXZ9 A0A146JYW4 C4B7V5 A0A0U2SQV8 A0A2H1V1Z3 A0A0E3VLQ3 A0A0P1IW02 A0A0E3VLQ7 A0A0E4B3V9 A0A2L1TG41 A0A2L1TG32 A0A0N7MCA7 U5NFR2 A0A386H8Y9 A0A1B3P5U6 A0A1S5XXP8 A0A1E1WPI5 A0A0B4ZWC0 Q6A1K2 A0A2L1TG21 A0A1B3B722 A0A3S2NXV2 R4JKH1 B5A7F1 A0A345BEY3 A0A3S2P9E5 A0A172QGF4 A0A075T622 R4JLY8 A0A1B2G2N4 A0A172QGF5 A0A2K8GKR8 R4JUT8 R4JSF7 A0A2W1BDT5 A0A2A4K9L1 A0A2H1V3Y7 A0A1B3P5M4 A0A1V1WBW6 A0A1B0GLR5 A0A0S1TPW8 A0A1V1WC75 A0A0S1TKX5 A0A1B3B742 A0A2K8GKS1
Pubmed
EMBL
NWSH01000229
PCG78177.1
PCG78178.1
KF768688
AIG51882.1
KY707205
+ More
AVF19647.1 NWSH01003963 PCG65612.1 MG546607 AYD42230.1 GCVX01000222 JAI18008.1 KY283629 AST36288.1 KY283630 AST36289.1 KX084473 ARO76428.1 KT588125 AOE48035.1 GDKB01000057 JAP38439.1 AB472105 BAH66322.1 BK005933 DAA05983.1 KM597232 AIT69909.1 GCVX01000221 JAI18009.1 KM655569 AIT72018.1 KU598912 ANZ03158.1 KM678333 AJE25906.1 MG816621 AXF48806.1 KM892407 AJF23822.1 KY283759 AST36418.1 GENK01000093 JAV45820.1 GEDO01000053 JAP88573.1 KY707214 AVF19656.1 KX585353 ARO70246.1 GDKB01000058 JAP38438.1 KX585333 ARO70226.1 KX655962 AOG12911.1 KQ459166 KPJ03567.1 KY283680 AST36339.1 KY225501 ARO70513.1 MH193982 AZB49428.1 KY283758 AST36417.1 ODYU01003236 SOQ41732.1 LN885103 CUQ99392.1 LN885127 CUQ99416.1 ODYU01001579 SOQ37997.1 LN885124 CUQ99413.1 GENK01000110 JAV45803.1 KR935758 ALM26250.1 KZ149944 PZC76857.1 KY399274 AQQ73505.1 KP975146 ALT31665.1 KY225491 ARO70503.1 KF487661 AII01059.1 AJ487482 CAD31853.1 KF487706 AII01104.1 LC002717 BAR43465.1 NWSH01000040 PCG80355.1 GEDO01000041 JAP88585.1 GEDO01000035 JAP88591.1 AB472104 BAH66321.1 KP975143 ALT31662.1 ODYU01000323 SOQ34863.1 LC002719 BAR43467.1 LN885105 CUQ99394.1 LC002739 BAR43487.1 LC002720 BAR43468.1 KY707219 AVF19661.1 KY707210 AVF19652.1 LN885132 CUQ99421.1 KC960456 AGY14567.1 MG546644 AYD42267.1 KX656010 AOG12959.1 KY399290 AQQ73521.1 GDQN01002308 JAT88746.1 KJ542666 AJD81550.1 AJ748327 CAG38113.1 KY707182 AVF19624.1 KT588107 AOE48017.1 RSAL01000029 RVE51760.1 JX982543 AGK90018.1 EU818704 ACF32963.1 MG816622 AXF48807.1 RSAL01000173 RVE45074.1 KT962963 AND95946.1 KF768666 KZ150109 AIG51860.1 PZC73368.1 JX982529 AGK90004.1 KU598902 ANZ03148.1 KT962962 AND95945.1 KX585334 ARO70227.1 JX982526 AGK90001.1 JX982540 AGK90015.1 PZC73369.1 PCG80352.1 ODYU01000356 SOQ34994.1 KX655969 AOG12918.1 GENK01000060 JAV45853.1 AJWK01030597 KR935738 ALM26230.1 GENK01000063 JAV45850.1 KR935710 ALM26202.1 KT588128 AOE48038.1 KX585336 ARO70229.1
AVF19647.1 NWSH01003963 PCG65612.1 MG546607 AYD42230.1 GCVX01000222 JAI18008.1 KY283629 AST36288.1 KY283630 AST36289.1 KX084473 ARO76428.1 KT588125 AOE48035.1 GDKB01000057 JAP38439.1 AB472105 BAH66322.1 BK005933 DAA05983.1 KM597232 AIT69909.1 GCVX01000221 JAI18009.1 KM655569 AIT72018.1 KU598912 ANZ03158.1 KM678333 AJE25906.1 MG816621 AXF48806.1 KM892407 AJF23822.1 KY283759 AST36418.1 GENK01000093 JAV45820.1 GEDO01000053 JAP88573.1 KY707214 AVF19656.1 KX585353 ARO70246.1 GDKB01000058 JAP38438.1 KX585333 ARO70226.1 KX655962 AOG12911.1 KQ459166 KPJ03567.1 KY283680 AST36339.1 KY225501 ARO70513.1 MH193982 AZB49428.1 KY283758 AST36417.1 ODYU01003236 SOQ41732.1 LN885103 CUQ99392.1 LN885127 CUQ99416.1 ODYU01001579 SOQ37997.1 LN885124 CUQ99413.1 GENK01000110 JAV45803.1 KR935758 ALM26250.1 KZ149944 PZC76857.1 KY399274 AQQ73505.1 KP975146 ALT31665.1 KY225491 ARO70503.1 KF487661 AII01059.1 AJ487482 CAD31853.1 KF487706 AII01104.1 LC002717 BAR43465.1 NWSH01000040 PCG80355.1 GEDO01000041 JAP88585.1 GEDO01000035 JAP88591.1 AB472104 BAH66321.1 KP975143 ALT31662.1 ODYU01000323 SOQ34863.1 LC002719 BAR43467.1 LN885105 CUQ99394.1 LC002739 BAR43487.1 LC002720 BAR43468.1 KY707219 AVF19661.1 KY707210 AVF19652.1 LN885132 CUQ99421.1 KC960456 AGY14567.1 MG546644 AYD42267.1 KX656010 AOG12959.1 KY399290 AQQ73521.1 GDQN01002308 JAT88746.1 KJ542666 AJD81550.1 AJ748327 CAG38113.1 KY707182 AVF19624.1 KT588107 AOE48017.1 RSAL01000029 RVE51760.1 JX982543 AGK90018.1 EU818704 ACF32963.1 MG816622 AXF48807.1 RSAL01000173 RVE45074.1 KT962963 AND95946.1 KF768666 KZ150109 AIG51860.1 PZC73368.1 JX982529 AGK90004.1 KU598902 ANZ03148.1 KT962962 AND95945.1 KX585334 ARO70227.1 JX982526 AGK90001.1 JX982540 AGK90015.1 PZC73369.1 PCG80352.1 ODYU01000356 SOQ34994.1 KX655969 AOG12918.1 GENK01000060 JAV45853.1 AJWK01030597 KR935738 ALM26230.1 GENK01000063 JAV45850.1 KR935710 ALM26202.1 KT588128 AOE48038.1 KX585336 ARO70229.1
Proteomes
Pfam
PF02949 7tm_6
Interpro
IPR004117
7tm6_olfct_rcpt
ProteinModelPortal
A0A2A4K1Q7
A0A2A4K2M3
A0A075T7Y9
A0A2L1TG45
A0A2A4J2I8
A0A386H9E1
+ More
A0A0K8TUP9 A0A223HD29 A0A223HD35 A0A1Y9TJS3 A0A1B3B766 A0A0V0J0N8 C4B7V6 A7E3G8 A0A097ITZ3 A0A0K8TU41 A0A097IYK3 A0A1B2G2P4 A0A0B5CSU1 A0A345BEY2 A0A0B5GQ56 A0A223HDH0 A0A1V1WC59 A0A146JVN1 A0A2L1TG60 A0A2K8GKT2 A0A0V0J2Q1 A0A2K8GKR7 A0A1B3P5N0 A0A0N1IA66 A0A223HD84 A0A2K8GL49 A0A3G6V789 A0A223HDF3 A0A2H1VMQ9 A0A0P1IW06 A0A0P1IVT0 A0A2H1VAZ7 A0A0P1J430 A0A1V1WBS9 A0A0S1TL39 A0A2W1BUH6 A0A1S5XXM4 A0A0U2QG96 A0A2K8GL39 A0A076E991 Q8MMH9 A0A076E9C7 A0A0E4B427 A0A2A4K7V7 A0A146JXZ9 A0A146JYW4 C4B7V5 A0A0U2SQV8 A0A2H1V1Z3 A0A0E3VLQ3 A0A0P1IW02 A0A0E3VLQ7 A0A0E4B3V9 A0A2L1TG41 A0A2L1TG32 A0A0N7MCA7 U5NFR2 A0A386H8Y9 A0A1B3P5U6 A0A1S5XXP8 A0A1E1WPI5 A0A0B4ZWC0 Q6A1K2 A0A2L1TG21 A0A1B3B722 A0A3S2NXV2 R4JKH1 B5A7F1 A0A345BEY3 A0A3S2P9E5 A0A172QGF4 A0A075T622 R4JLY8 A0A1B2G2N4 A0A172QGF5 A0A2K8GKR8 R4JUT8 R4JSF7 A0A2W1BDT5 A0A2A4K9L1 A0A2H1V3Y7 A0A1B3P5M4 A0A1V1WBW6 A0A1B0GLR5 A0A0S1TPW8 A0A1V1WC75 A0A0S1TKX5 A0A1B3B742 A0A2K8GKS1
A0A0K8TUP9 A0A223HD29 A0A223HD35 A0A1Y9TJS3 A0A1B3B766 A0A0V0J0N8 C4B7V6 A7E3G8 A0A097ITZ3 A0A0K8TU41 A0A097IYK3 A0A1B2G2P4 A0A0B5CSU1 A0A345BEY2 A0A0B5GQ56 A0A223HDH0 A0A1V1WC59 A0A146JVN1 A0A2L1TG60 A0A2K8GKT2 A0A0V0J2Q1 A0A2K8GKR7 A0A1B3P5N0 A0A0N1IA66 A0A223HD84 A0A2K8GL49 A0A3G6V789 A0A223HDF3 A0A2H1VMQ9 A0A0P1IW06 A0A0P1IVT0 A0A2H1VAZ7 A0A0P1J430 A0A1V1WBS9 A0A0S1TL39 A0A2W1BUH6 A0A1S5XXM4 A0A0U2QG96 A0A2K8GL39 A0A076E991 Q8MMH9 A0A076E9C7 A0A0E4B427 A0A2A4K7V7 A0A146JXZ9 A0A146JYW4 C4B7V5 A0A0U2SQV8 A0A2H1V1Z3 A0A0E3VLQ3 A0A0P1IW02 A0A0E3VLQ7 A0A0E4B3V9 A0A2L1TG41 A0A2L1TG32 A0A0N7MCA7 U5NFR2 A0A386H8Y9 A0A1B3P5U6 A0A1S5XXP8 A0A1E1WPI5 A0A0B4ZWC0 Q6A1K2 A0A2L1TG21 A0A1B3B722 A0A3S2NXV2 R4JKH1 B5A7F1 A0A345BEY3 A0A3S2P9E5 A0A172QGF4 A0A075T622 R4JLY8 A0A1B2G2N4 A0A172QGF5 A0A2K8GKR8 R4JUT8 R4JSF7 A0A2W1BDT5 A0A2A4K9L1 A0A2H1V3Y7 A0A1B3P5M4 A0A1V1WBW6 A0A1B0GLR5 A0A0S1TPW8 A0A1V1WC75 A0A0S1TKX5 A0A1B3B742 A0A2K8GKS1
PDB
6C70
E-value=0.00241024,
Score=90
Ontologies
GO
PANTHER
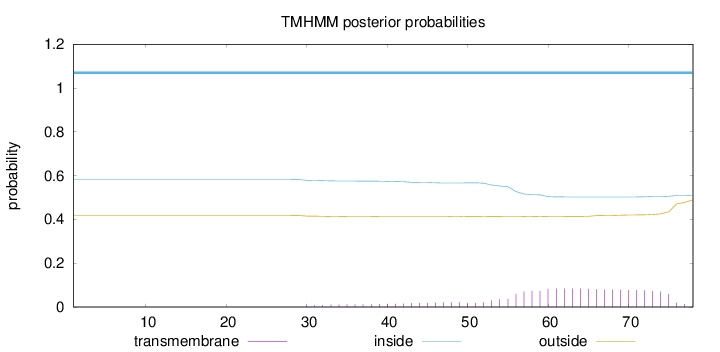
Topology
Subcellular location
Cell membrane
Length:
78
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.01445
Exp number, first 60 AAs:
0.81278
Total prob of N-in:
0.58221
inside
1 - 78
Population Genetic Test Statistics
Pi
377.228985
Theta
197.377956
Tajima's D
2.868006
CLR
0.590575
CSRT
0.972551372431378
Interpretation
Uncertain
