Gene
KWMTBOMO10739 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA008408
Annotation
PREDICTED:_kinesin_light_chain_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.811 Nuclear Reliability : 2.079
Sequence
CDS
ATGACGGCTCTGGCATCTCACCTTCAGTCCGTCGAGGCAGAGAAGCAAAAACTCCGCACGCAGGTCCGCAGGCTGTGCCAAGAGAACGCCTGGCTGAGGGACGAGCTGGCTGCAGCTCAGCAGCACCTGCAGGCTTCCGAACAACGCGTAGCACAGCTGGAGGAGGAGAATAAGCATTTGGAGTTCATGGCTTCCATACGTAAATACGACAGCGACATAAACGAAGAGTCGGACAGTAACCGTACAGGTCAGGGCGAGAAGAAACGCGAGGACCCGATGGTCGAGCTGTTCCCTGACGATGACCACGACGACAACAGGAATAATAAATCGATGTCGCCGACGCCGCCGATGCACTTCGCCCAGCAAGTGAACGCCGGGTACGAGATCCCGGCCCGCCTGCGCACGCTCCACAACCTGGTCATACAGTACGCGTCGCAGGGCCGGTACGAGGTCGCCGTGCCCCTCTGCAAGCAAGCACTCGAAGACCTGGAGAAGACTTCGGGCCACGACCATCCCGATGTCGCCACCATGCTGAACATACTGGCTCTTGTTTACAGAGACCAAAACAAATACAAGGAAGCCGCCAACCTGCTGAACGACGCTCTGTCCATCCGGGAGAAGACCCTCGGCGAGAACCACCCCGCCGTGGCCGCCACGCTCAACAACCTCGCCGTGCTCTACGGGAAGCGAGGCAAGTACCGCGAGGCGGAGCCGCTGTGCAAGCGCGCGCTGTGCATCCGCGAGGCCGTGCTGGGCCGCGACCACCCCGACGTCGCCAAGCAGCTCAACAACCTGGCGCTGCTCTGCCAGAACCAGAACAAGTACGAGGAGGTGGAGCAGTACTACCAGCGCGCGCTCGAGATCTACGAGAGCAAGCTCGGACCCGACGACCCCAACGTTGCCAAGACCAAGAACAACCTCGCCTCCTGCTATCTCAAACAGGGCAAATACAAGGAAGCCGAGACTCTCTACAAGCAGGTGTTGACGCGGGCTCATGAACGAGAATTCGGAACCATTGACGAATATATAGCATCGTTAAAAAAACCTAAAAAATAA
Protein
MTALASHLQSVEAEKQKLRTQVRRLCQENAWLRDELAAAQQHLQASEQRVAQLEEENKHLEFMASIRKYDSDINEESDSNRTGQGEKKREDPMVELFPDDDHDDNRNNKSMSPTPPMHFAQQVNAGYEIPARLRTLHNLVIQYASQGRYEVAVPLCKQALEDLEKTSGHDHPDVATMLNILALVYRDQNKYKEAANLLNDALSIREKTLGENHPAVAATLNNLAVLYGKRGKYREAEPLCKRALCIREAVLGRDHPDVAKQLNNLALLCQNQNKYEEVEQYYQRALEIYESKLGPDDPNVAKTKNNLASCYLKQGKYKEAETLYKQVLTRAHEREFGTIDEYIASLKKPKK
Summary
Uniprot
A0A2A4K2E6
A0A3S2M0B0
S4PWN8
A0A1E1WLV0
H9JFW1
A0A2A4K338
+ More
A0A2A4K1P4 A0A2A4K2Q4 A0A2A4K2P3 A0A1E1WDG1 A0A0N1IAJ4 A0A212EUK1 A0A2J7PW86 A0A2P8YJA6 A0A224XP90 A0A0V0G8F3 A0A0N7Z9L6 A0A069DVL8 A0A1B6MPI3 T1HLE6 A0A067QTL0 A0A1B6GZU1 A0A1B6F1D6 A0A1B6H419 A0A0K8SGW0 A0A195CAF0 A0A0K8SGV5 A0A0K8SGV9 A0A0K8SGV6 A0A0K8SHF4 A0A0K8SGV4 A0A2H8TJH5 A0A2S2QK68 K7IU41 J9KB84 A0A3Q0ILD3 U5EKC3 E9I8X3 D2A3J4 A0A0J7KQZ1 A0A2S2NYH9 A0A158NJI9 A0A1L8E558 F4X3W3 A0A0C9QXD8 A0A1B0CB71 A0A0C9RPH6 A0A026WZS5 A0A1J1IID1 A0A423TT64 A0A1Y1LDA8 A0A3L8DRX9 T1DI28 T1E2K3 A0A0P5ME20 A0A0P5KBY2 A0A0P5GLT8 A0A0P5D342 A0A0P5I0S3 A0A0P6F3T3 A0A0P6F9D8 A0A0P5W301 A0A1Q3FG08 A0A0P5EHA9 A0A0P5NBG1 A0A0P6E5B1 A0A0P5EFC8 A0A0P4YTP6 B0X9A9 A0A0P5Y328 A0A0P5N7N3 A0A0P5SZX0 A0A0N8C420 A0A0P5HT85 A0A0P5RYL8 A0A0P5RL84 A0A1Y1LFZ8 A0A0P5NX59 A0A0P5KKQ0 A0A0P5EN09 A0A0N8DR52 A0A0P5C1R4 A0A0N8E9G6 A0A0P5BQK7 A0A0P5THZ0 E9G6H5 A0A023ETF8 A0A0P5MZN5 Q16LZ3 A0A0P5UCK0 A0A1Y1LEF3 A0A0P5WNL3 A0A0P4YX51 A0A0P5I599 A0A0P6F6X0 A0A0P5E1A6 A0A0P5KNN2 A0A0P5ZNU0 A0A0N8DCX4 A0A0P5DXJ7
A0A2A4K1P4 A0A2A4K2Q4 A0A2A4K2P3 A0A1E1WDG1 A0A0N1IAJ4 A0A212EUK1 A0A2J7PW86 A0A2P8YJA6 A0A224XP90 A0A0V0G8F3 A0A0N7Z9L6 A0A069DVL8 A0A1B6MPI3 T1HLE6 A0A067QTL0 A0A1B6GZU1 A0A1B6F1D6 A0A1B6H419 A0A0K8SGW0 A0A195CAF0 A0A0K8SGV5 A0A0K8SGV9 A0A0K8SGV6 A0A0K8SHF4 A0A0K8SGV4 A0A2H8TJH5 A0A2S2QK68 K7IU41 J9KB84 A0A3Q0ILD3 U5EKC3 E9I8X3 D2A3J4 A0A0J7KQZ1 A0A2S2NYH9 A0A158NJI9 A0A1L8E558 F4X3W3 A0A0C9QXD8 A0A1B0CB71 A0A0C9RPH6 A0A026WZS5 A0A1J1IID1 A0A423TT64 A0A1Y1LDA8 A0A3L8DRX9 T1DI28 T1E2K3 A0A0P5ME20 A0A0P5KBY2 A0A0P5GLT8 A0A0P5D342 A0A0P5I0S3 A0A0P6F3T3 A0A0P6F9D8 A0A0P5W301 A0A1Q3FG08 A0A0P5EHA9 A0A0P5NBG1 A0A0P6E5B1 A0A0P5EFC8 A0A0P4YTP6 B0X9A9 A0A0P5Y328 A0A0P5N7N3 A0A0P5SZX0 A0A0N8C420 A0A0P5HT85 A0A0P5RYL8 A0A0P5RL84 A0A1Y1LFZ8 A0A0P5NX59 A0A0P5KKQ0 A0A0P5EN09 A0A0N8DR52 A0A0P5C1R4 A0A0N8E9G6 A0A0P5BQK7 A0A0P5THZ0 E9G6H5 A0A023ETF8 A0A0P5MZN5 Q16LZ3 A0A0P5UCK0 A0A1Y1LEF3 A0A0P5WNL3 A0A0P4YX51 A0A0P5I599 A0A0P6F6X0 A0A0P5E1A6 A0A0P5KNN2 A0A0P5ZNU0 A0A0N8DCX4 A0A0P5DXJ7
Pubmed
EMBL
NWSH01000229
PCG78186.1
RSAL01000088
RVE48184.1
GAIX01006463
JAA86097.1
+ More
GDQN01003228 JAT87826.1 BABH01001529 BABH01001530 PCG78193.1 PCG78191.1 PCG78184.1 PCG78198.1 GDQN01006030 GDQN01005027 JAT85024.1 JAT86027.1 KQ460326 KPJ15665.1 AGBW02012362 OWR45163.1 NEVH01020936 PNF20602.1 PYGN01000554 PSN44341.1 GFTR01006176 JAW10250.1 GECL01001756 JAP04368.1 GDKW01000203 JAI56392.1 GBGD01000949 JAC87940.1 GEBQ01002165 JAT37812.1 ACPB03017689 ACPB03017690 KK852969 KDR13110.1 GECZ01001815 JAS67954.1 GECZ01025707 JAS44062.1 GECZ01000382 JAS69387.1 GBRD01013300 JAG52526.1 KQ978081 KYM97093.1 GBRD01013304 GBRD01013299 JAG52522.1 GBRD01013298 JAG52528.1 GBRD01013305 GBRD01013301 JAG52521.1 GBRD01013302 JAG52524.1 GBRD01013306 GBRD01013303 JAG52523.1 GFXV01002492 MBW14297.1 GGMS01008921 MBY78124.1 AAZX01002657 ABLF02037027 GANO01001864 JAB58007.1 GL761662 EFZ22996.1 KQ971338 EFA02324.2 LBMM01004197 KMQ92711.1 GGMR01009616 MBY22235.1 ADTU01017944 ADTU01017945 GFDF01000204 JAV13880.1 GL888624 EGI58910.1 GBYB01000345 JAG70112.1 AJWK01004867 AJWK01004868 GBYB01010340 JAG80107.1 KK107063 EZA61246.1 CVRI01000050 CRK99284.1 QCYY01001210 ROT79649.1 GEZM01062236 JAV69935.1 QOIP01000005 RLU23164.1 GALA01001186 JAA93666.1 GALA01001185 JAA93667.1 GDIQ01165177 JAK86548.1 GDIQ01192752 JAK58973.1 GDIQ01239446 JAK12279.1 GDIP01161938 JAJ61464.1 GDIQ01220054 JAK31671.1 GDIQ01068254 JAN26483.1 GDIQ01053563 JAN41174.1 GDIP01092484 JAM11231.1 GFDL01008555 JAV26490.1 GDIP01148490 JAJ74912.1 GDIQ01167111 JAK84614.1 GDIQ01068253 JAN26484.1 GDIP01161029 JAJ62373.1 GDIP01222829 JAJ00573.1 DS232529 EDS43041.1 GDIP01063702 GDIQ01066757 JAM40013.1 JAN27980.1 GDIQ01149874 JAL01852.1 GDIP01132986 JAL70728.1 GDIQ01116899 JAL34827.1 GDIQ01225340 JAK26385.1 GDIQ01094593 JAL57133.1 GDIQ01100797 JAL50929.1 GEZM01062234 JAV69937.1 GDIQ01138685 JAL13041.1 GDIQ01182766 JAK68959.1 GDIP01145692 JAJ77710.1 GDIP01009031 JAM94684.1 GDIP01176784 JAJ46618.1 GDIQ01048740 JAN45997.1 GDIP01186034 JAJ37368.1 GDIP01131749 JAL71965.1 GL732533 EFX84937.1 GAPW01001015 JAC12583.1 GDIQ01171308 JAK80417.1 CH477886 EAT35354.1 GDIP01114994 JAL88720.1 GEZM01062235 JAV69936.1 GDIP01096448 JAM07267.1 GDIP01221789 JAJ01613.1 GDIQ01220055 JAK31670.1 GDIQ01051920 JAN42817.1 GDIP01166341 JAJ57061.1 GDIQ01182767 JAK68958.1 GDIP01042140 JAM61575.1 GDIP01046055 JAM57660.1 GDIP01151034 JAJ72368.1
GDQN01003228 JAT87826.1 BABH01001529 BABH01001530 PCG78193.1 PCG78191.1 PCG78184.1 PCG78198.1 GDQN01006030 GDQN01005027 JAT85024.1 JAT86027.1 KQ460326 KPJ15665.1 AGBW02012362 OWR45163.1 NEVH01020936 PNF20602.1 PYGN01000554 PSN44341.1 GFTR01006176 JAW10250.1 GECL01001756 JAP04368.1 GDKW01000203 JAI56392.1 GBGD01000949 JAC87940.1 GEBQ01002165 JAT37812.1 ACPB03017689 ACPB03017690 KK852969 KDR13110.1 GECZ01001815 JAS67954.1 GECZ01025707 JAS44062.1 GECZ01000382 JAS69387.1 GBRD01013300 JAG52526.1 KQ978081 KYM97093.1 GBRD01013304 GBRD01013299 JAG52522.1 GBRD01013298 JAG52528.1 GBRD01013305 GBRD01013301 JAG52521.1 GBRD01013302 JAG52524.1 GBRD01013306 GBRD01013303 JAG52523.1 GFXV01002492 MBW14297.1 GGMS01008921 MBY78124.1 AAZX01002657 ABLF02037027 GANO01001864 JAB58007.1 GL761662 EFZ22996.1 KQ971338 EFA02324.2 LBMM01004197 KMQ92711.1 GGMR01009616 MBY22235.1 ADTU01017944 ADTU01017945 GFDF01000204 JAV13880.1 GL888624 EGI58910.1 GBYB01000345 JAG70112.1 AJWK01004867 AJWK01004868 GBYB01010340 JAG80107.1 KK107063 EZA61246.1 CVRI01000050 CRK99284.1 QCYY01001210 ROT79649.1 GEZM01062236 JAV69935.1 QOIP01000005 RLU23164.1 GALA01001186 JAA93666.1 GALA01001185 JAA93667.1 GDIQ01165177 JAK86548.1 GDIQ01192752 JAK58973.1 GDIQ01239446 JAK12279.1 GDIP01161938 JAJ61464.1 GDIQ01220054 JAK31671.1 GDIQ01068254 JAN26483.1 GDIQ01053563 JAN41174.1 GDIP01092484 JAM11231.1 GFDL01008555 JAV26490.1 GDIP01148490 JAJ74912.1 GDIQ01167111 JAK84614.1 GDIQ01068253 JAN26484.1 GDIP01161029 JAJ62373.1 GDIP01222829 JAJ00573.1 DS232529 EDS43041.1 GDIP01063702 GDIQ01066757 JAM40013.1 JAN27980.1 GDIQ01149874 JAL01852.1 GDIP01132986 JAL70728.1 GDIQ01116899 JAL34827.1 GDIQ01225340 JAK26385.1 GDIQ01094593 JAL57133.1 GDIQ01100797 JAL50929.1 GEZM01062234 JAV69937.1 GDIQ01138685 JAL13041.1 GDIQ01182766 JAK68959.1 GDIP01145692 JAJ77710.1 GDIP01009031 JAM94684.1 GDIP01176784 JAJ46618.1 GDIQ01048740 JAN45997.1 GDIP01186034 JAJ37368.1 GDIP01131749 JAL71965.1 GL732533 EFX84937.1 GAPW01001015 JAC12583.1 GDIQ01171308 JAK80417.1 CH477886 EAT35354.1 GDIP01114994 JAL88720.1 GEZM01062235 JAV69936.1 GDIP01096448 JAM07267.1 GDIP01221789 JAJ01613.1 GDIQ01220055 JAK31670.1 GDIQ01051920 JAN42817.1 GDIP01166341 JAJ57061.1 GDIQ01182767 JAK68958.1 GDIP01042140 JAM61575.1 GDIP01046055 JAM57660.1 GDIP01151034 JAJ72368.1
Proteomes
UP000218220
UP000283053
UP000005204
UP000053240
UP000007151
UP000235965
+ More
UP000245037 UP000015103 UP000027135 UP000078542 UP000002358 UP000007819 UP000079169 UP000007266 UP000036403 UP000005205 UP000007755 UP000092461 UP000053097 UP000183832 UP000283509 UP000279307 UP000002320 UP000000305 UP000008820
UP000245037 UP000015103 UP000027135 UP000078542 UP000002358 UP000007819 UP000079169 UP000007266 UP000036403 UP000005205 UP000007755 UP000092461 UP000053097 UP000183832 UP000283509 UP000279307 UP000002320 UP000000305 UP000008820
PRIDE
Interpro
SUPFAM
SSF48452
SSF48452
Gene 3D
ProteinModelPortal
A0A2A4K2E6
A0A3S2M0B0
S4PWN8
A0A1E1WLV0
H9JFW1
A0A2A4K338
+ More
A0A2A4K1P4 A0A2A4K2Q4 A0A2A4K2P3 A0A1E1WDG1 A0A0N1IAJ4 A0A212EUK1 A0A2J7PW86 A0A2P8YJA6 A0A224XP90 A0A0V0G8F3 A0A0N7Z9L6 A0A069DVL8 A0A1B6MPI3 T1HLE6 A0A067QTL0 A0A1B6GZU1 A0A1B6F1D6 A0A1B6H419 A0A0K8SGW0 A0A195CAF0 A0A0K8SGV5 A0A0K8SGV9 A0A0K8SGV6 A0A0K8SHF4 A0A0K8SGV4 A0A2H8TJH5 A0A2S2QK68 K7IU41 J9KB84 A0A3Q0ILD3 U5EKC3 E9I8X3 D2A3J4 A0A0J7KQZ1 A0A2S2NYH9 A0A158NJI9 A0A1L8E558 F4X3W3 A0A0C9QXD8 A0A1B0CB71 A0A0C9RPH6 A0A026WZS5 A0A1J1IID1 A0A423TT64 A0A1Y1LDA8 A0A3L8DRX9 T1DI28 T1E2K3 A0A0P5ME20 A0A0P5KBY2 A0A0P5GLT8 A0A0P5D342 A0A0P5I0S3 A0A0P6F3T3 A0A0P6F9D8 A0A0P5W301 A0A1Q3FG08 A0A0P5EHA9 A0A0P5NBG1 A0A0P6E5B1 A0A0P5EFC8 A0A0P4YTP6 B0X9A9 A0A0P5Y328 A0A0P5N7N3 A0A0P5SZX0 A0A0N8C420 A0A0P5HT85 A0A0P5RYL8 A0A0P5RL84 A0A1Y1LFZ8 A0A0P5NX59 A0A0P5KKQ0 A0A0P5EN09 A0A0N8DR52 A0A0P5C1R4 A0A0N8E9G6 A0A0P5BQK7 A0A0P5THZ0 E9G6H5 A0A023ETF8 A0A0P5MZN5 Q16LZ3 A0A0P5UCK0 A0A1Y1LEF3 A0A0P5WNL3 A0A0P4YX51 A0A0P5I599 A0A0P6F6X0 A0A0P5E1A6 A0A0P5KNN2 A0A0P5ZNU0 A0A0N8DCX4 A0A0P5DXJ7
A0A2A4K1P4 A0A2A4K2Q4 A0A2A4K2P3 A0A1E1WDG1 A0A0N1IAJ4 A0A212EUK1 A0A2J7PW86 A0A2P8YJA6 A0A224XP90 A0A0V0G8F3 A0A0N7Z9L6 A0A069DVL8 A0A1B6MPI3 T1HLE6 A0A067QTL0 A0A1B6GZU1 A0A1B6F1D6 A0A1B6H419 A0A0K8SGW0 A0A195CAF0 A0A0K8SGV5 A0A0K8SGV9 A0A0K8SGV6 A0A0K8SHF4 A0A0K8SGV4 A0A2H8TJH5 A0A2S2QK68 K7IU41 J9KB84 A0A3Q0ILD3 U5EKC3 E9I8X3 D2A3J4 A0A0J7KQZ1 A0A2S2NYH9 A0A158NJI9 A0A1L8E558 F4X3W3 A0A0C9QXD8 A0A1B0CB71 A0A0C9RPH6 A0A026WZS5 A0A1J1IID1 A0A423TT64 A0A1Y1LDA8 A0A3L8DRX9 T1DI28 T1E2K3 A0A0P5ME20 A0A0P5KBY2 A0A0P5GLT8 A0A0P5D342 A0A0P5I0S3 A0A0P6F3T3 A0A0P6F9D8 A0A0P5W301 A0A1Q3FG08 A0A0P5EHA9 A0A0P5NBG1 A0A0P6E5B1 A0A0P5EFC8 A0A0P4YTP6 B0X9A9 A0A0P5Y328 A0A0P5N7N3 A0A0P5SZX0 A0A0N8C420 A0A0P5HT85 A0A0P5RYL8 A0A0P5RL84 A0A1Y1LFZ8 A0A0P5NX59 A0A0P5KKQ0 A0A0P5EN09 A0A0N8DR52 A0A0P5C1R4 A0A0N8E9G6 A0A0P5BQK7 A0A0P5THZ0 E9G6H5 A0A023ETF8 A0A0P5MZN5 Q16LZ3 A0A0P5UCK0 A0A1Y1LEF3 A0A0P5WNL3 A0A0P4YX51 A0A0P5I599 A0A0P6F6X0 A0A0P5E1A6 A0A0P5KNN2 A0A0P5ZNU0 A0A0N8DCX4 A0A0P5DXJ7
PDB
3NF1
E-value=6.23233e-91,
Score=852
Ontologies
GO
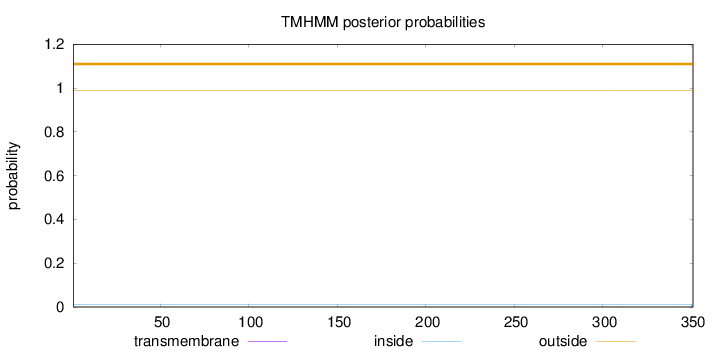
Topology
Length:
351
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00127
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01230
outside
1 - 351
Population Genetic Test Statistics
Pi
183.008603
Theta
164.123083
Tajima's D
0.458
CLR
0
CSRT
0.501424928753562
Interpretation
Uncertain
