Gene
KWMTBOMO10719
Pre Gene Modal
BGIBMGA008281
Annotation
PREDICTED:_ovochymase-2-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.975 PlasmaMembrane Reliability : 2.638
Sequence
CDS
ATGAACGTAAAATACGGAATACTTTTTGTGGCGCTCTTTGCGGCTGCCTGGGCCCTCCCCAAGCCTGAAGATGACATGTCCATCTTCTATGAGCATGTGGACCGGAACGCTCGTATTGTGGGGGGTAGCCAGGCAGCTCAGGGCAGCCACCCCCACATGGTAGCGATGACTAATGGTATCTTCATCAGGAGCTTTGTCTGCGGAGGCTCAGTGCTCACCGCACGCACGGTGCTCACCGCTGCTCATTGCATTGCCGCTGTATTCATATTTGGAAGTCTCTCTGGAAATCTTCGTCTGACAGTCGGTACAAACCAATGGAACTCTGGTGGTACCCTCCACGCTGTCTCCAGAAACATCACCCACCCTAACTACGTGTCCAATACTATCAAGAACGACTTGGGTATTCTCATAACTTCCTCGAACATCGTCTTCAACGACCGCGTCAGGCCTATCTCTTTGAGTTTTGACTACGTACCTGGAGGCGTCCCTGTCAGGGTTGCTGGATGGGGAAGAGTTAGGGCTAACGGTGCTCTCTCCACGAATCTTCTGGAGATCAATGTGAGAACCATTGATGCTCAGACTTGCGTGAGGACTGCGGCCCAGGCGGCCATTGATCTTAACATGAGGGCTCCTCCTGTCGAACCTCACATCGAGCTCTGCACATTCCACGCAGAAGGAACTGGTACTTGCAATGGTGACAGTGGCAGCGCATTGGCTCGGACCGACAGCGGTCTCCAAGTCGGCATCGTGTCGTGGGGCTTCCCTTGCGCGCTGGGAGCTCCTGACATGTTCGTCAGAGTCAGCGCCTTCCGGTCCTGGCTGCAACAAAACATAGCTAATTAA
Protein
MNVKYGILFVALFAAAWALPKPEDDMSIFYEHVDRNARIVGGSQAAQGSHPHMVAMTNGIFIRSFVCGGSVLTARTVLTAAHCIAAVFIFGSLSGNLRLTVGTNQWNSGGTLHAVSRNITHPNYVSNTIKNDLGILITSSNIVFNDRVRPISLSFDYVPGGVPVRVAGWGRVRANGALSTNLLEINVRTIDAQTCVRTAAQAAIDLNMRAPPVEPHIELCTFHAEGTGTCNGDSGSALARTDSGLQVGIVSWGFPCALGAPDMFVRVSAFRSWLQQNIAN
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9JFI4
H9BVM5
H9JFI3
H9JFI2
B6CME8
B4X984
+ More
A4F5H5 A0A2A4J186 C9W8H0 A0A2H1VW00 A0A0X9GKP8 A0A2A4J150 A5CG73 A5CG74 A0A194PCH8 A0A2H1WEC1 H9JFI1 E7D011 A0A2H1VHU4 I6SW54 A0A1E1WV71 A5CG72 A0A0N1PHG9 A0A2H1VG38 A0A1E1W396 A0A3S2M0D3 A0A2H1VIU2 A0A212F2F5 A0A2H1VG36 A0A0N0PD42 A0A2H1VAF3 A0A194PCP7 A0A212FHN1 A0A212F2H0 A0A0L7LAZ5 A0A2H1VHM1 A0A212FIQ7 A0A0N0PDI4 A0A194PIR8 A0A2H1VG24 A0A0L7LGT4 A0A2H1WNQ2 A0A3S2LJS9 A0A2H1WSA0 H9JFE5 A0A212FHN6 A0A1E1WG09 A0A2A4JX80 A0A0L7L2A5 I6TEX4 A0A2W1BP48 Q3ZJD2 A0A194PE67 A0A212F7H7 A0A2H1X2T2 A0A2A4J0U4 A0A2W1BHD3 A0A2H1X2X6 A0A0N0PD79 A0A1Q3FSI0 A0A212F7I7 A0A154P6Q3 E1ZWT5 A0A1B0GNP3 Q9XY45 B0WW46 A0A1Q3FHD4 A0A1Q3FNV0 A0A3S2NS83 K7IRM7 A0A023EMN4 B0WLB8 Q8T639 A0A026WVM2 A0A087ZQ85 O97099 A0A182I4T2 Q7PF16 E2ACM3 W5JMT6 B0WHA0 A0A0M9A2K3 A0A182X1C1 A0A023EMU9 A0A1W7R7W0 H9JWP8 T1DGK1 A8CA02 A0A2A3EKL4 A0A0J7KPH6 A0A182KLF3 A0A1Y1MLX5 A0A232EZ15 A0A212F633 A0A0J7K549 A0A2M4CW71 A0A1S4FE82 A0A182UFJ4 A0A182V9J9
A4F5H5 A0A2A4J186 C9W8H0 A0A2H1VW00 A0A0X9GKP8 A0A2A4J150 A5CG73 A5CG74 A0A194PCH8 A0A2H1WEC1 H9JFI1 E7D011 A0A2H1VHU4 I6SW54 A0A1E1WV71 A5CG72 A0A0N1PHG9 A0A2H1VG38 A0A1E1W396 A0A3S2M0D3 A0A2H1VIU2 A0A212F2F5 A0A2H1VG36 A0A0N0PD42 A0A2H1VAF3 A0A194PCP7 A0A212FHN1 A0A212F2H0 A0A0L7LAZ5 A0A2H1VHM1 A0A212FIQ7 A0A0N0PDI4 A0A194PIR8 A0A2H1VG24 A0A0L7LGT4 A0A2H1WNQ2 A0A3S2LJS9 A0A2H1WSA0 H9JFE5 A0A212FHN6 A0A1E1WG09 A0A2A4JX80 A0A0L7L2A5 I6TEX4 A0A2W1BP48 Q3ZJD2 A0A194PE67 A0A212F7H7 A0A2H1X2T2 A0A2A4J0U4 A0A2W1BHD3 A0A2H1X2X6 A0A0N0PD79 A0A1Q3FSI0 A0A212F7I7 A0A154P6Q3 E1ZWT5 A0A1B0GNP3 Q9XY45 B0WW46 A0A1Q3FHD4 A0A1Q3FNV0 A0A3S2NS83 K7IRM7 A0A023EMN4 B0WLB8 Q8T639 A0A026WVM2 A0A087ZQ85 O97099 A0A182I4T2 Q7PF16 E2ACM3 W5JMT6 B0WHA0 A0A0M9A2K3 A0A182X1C1 A0A023EMU9 A0A1W7R7W0 H9JWP8 T1DGK1 A8CA02 A0A2A3EKL4 A0A0J7KPH6 A0A182KLF3 A0A1Y1MLX5 A0A232EZ15 A0A212F633 A0A0J7K549 A0A2M4CW71 A0A1S4FE82 A0A182UFJ4 A0A182V9J9
Pubmed
EMBL
BABH01035039
BABH01035040
JQ081296
AFD99127.1
BABH01035036
BABH01035037
+ More
EU325548 KZ150045 ACB54938.1 PZC74508.1 EF531620 ABR88231.1 AM419170 CAL92020.1 NWSH01004418 PCG65143.1 FJ205415 ACR15983.2 ODYU01004616 SOQ44682.1 KP690082 ODYU01004615 ALO61084.1 SOQ44680.1 PCG65142.1 AM690448 CAM84318.1 AM690449 CAM84319.1 KQ459606 KPI90986.1 ODYU01008037 SOQ51306.1 BABH01035035 HM990181 ADT80829.1 ODYU01002608 SOQ40331.1 JQ904145 AFM77775.1 GDQN01000207 JAT90847.1 AM690447 CAM84317.1 KQ460326 KPJ15683.1 ODYU01002379 SOQ39797.1 GDQN01009632 JAT81422.1 RSAL01000088 RVE48198.1 SOQ40332.1 AGBW02010744 OWR47925.1 SOQ39799.1 KPJ15684.1 ODYU01001513 SOQ37833.1 KPI90987.1 AGBW02008493 OWR53226.1 OWR47924.1 JTDY01001984 KOB72401.1 SOQ40333.1 AGBW02008353 OWR53602.1 KPJ15682.1 KPI90985.1 SOQ39798.1 JTDY01001141 KOB74753.1 ODYU01009947 SOQ54703.1 RVE48200.1 ODYU01010616 SOQ55846.1 BABH01035041 OWR53227.1 GDQN01005253 JAT85801.1 NWSH01000498 PCG76020.1 JTDY01003392 KOB69628.1 JQ904144 AFM77774.1 PZC74510.1 AY820894 AAX35812.1 KPI90984.1 AGBW02009869 OWR49695.1 ODYU01013016 SOQ59625.1 PCG65144.1 PZC74509.1 SOQ59627.1 KPJ15681.1 GFDL01004672 JAV30373.1 OWR49696.1 KQ434827 KZC07503.1 GL434905 EFN74332.1 AJVK01029631 AF053903 AAD21823.1 DS232137 EDS35878.1 GFDL01008064 JAV26981.1 GFDL01005774 JAV29271.1 RSAL01000269 RVE43195.1 GAPW01003508 JAC10090.1 DS232962 DS231983 EDS27062.1 EDS30402.1 AF487334 AAL93243.1 KK107109 QOIP01000008 EZA59134.1 RLU19794.1 AF051780 AAD17493.1 APCN01002347 AAAB01008807 EAA45498.1 GL438568 EFN68767.1 ADMH02001109 ETN64084.1 DS231933 EDS27558.1 KQ435756 KOX75917.1 GAPW01003418 JAC10180.1 GEHC01000393 JAV47252.1 BABH01018411 GAMD01002746 JAA98844.1 EU035826 ABV44728.1 KZ288229 PBC31726.1 LBMM01004589 KMQ92263.1 GEZM01027546 JAV86682.1 NNAY01001573 OXU23549.1 AGBW02010081 OWR49201.1 LBMM01013914 KMQ85419.1 GGFL01005406 MBW69584.1
EU325548 KZ150045 ACB54938.1 PZC74508.1 EF531620 ABR88231.1 AM419170 CAL92020.1 NWSH01004418 PCG65143.1 FJ205415 ACR15983.2 ODYU01004616 SOQ44682.1 KP690082 ODYU01004615 ALO61084.1 SOQ44680.1 PCG65142.1 AM690448 CAM84318.1 AM690449 CAM84319.1 KQ459606 KPI90986.1 ODYU01008037 SOQ51306.1 BABH01035035 HM990181 ADT80829.1 ODYU01002608 SOQ40331.1 JQ904145 AFM77775.1 GDQN01000207 JAT90847.1 AM690447 CAM84317.1 KQ460326 KPJ15683.1 ODYU01002379 SOQ39797.1 GDQN01009632 JAT81422.1 RSAL01000088 RVE48198.1 SOQ40332.1 AGBW02010744 OWR47925.1 SOQ39799.1 KPJ15684.1 ODYU01001513 SOQ37833.1 KPI90987.1 AGBW02008493 OWR53226.1 OWR47924.1 JTDY01001984 KOB72401.1 SOQ40333.1 AGBW02008353 OWR53602.1 KPJ15682.1 KPI90985.1 SOQ39798.1 JTDY01001141 KOB74753.1 ODYU01009947 SOQ54703.1 RVE48200.1 ODYU01010616 SOQ55846.1 BABH01035041 OWR53227.1 GDQN01005253 JAT85801.1 NWSH01000498 PCG76020.1 JTDY01003392 KOB69628.1 JQ904144 AFM77774.1 PZC74510.1 AY820894 AAX35812.1 KPI90984.1 AGBW02009869 OWR49695.1 ODYU01013016 SOQ59625.1 PCG65144.1 PZC74509.1 SOQ59627.1 KPJ15681.1 GFDL01004672 JAV30373.1 OWR49696.1 KQ434827 KZC07503.1 GL434905 EFN74332.1 AJVK01029631 AF053903 AAD21823.1 DS232137 EDS35878.1 GFDL01008064 JAV26981.1 GFDL01005774 JAV29271.1 RSAL01000269 RVE43195.1 GAPW01003508 JAC10090.1 DS232962 DS231983 EDS27062.1 EDS30402.1 AF487334 AAL93243.1 KK107109 QOIP01000008 EZA59134.1 RLU19794.1 AF051780 AAD17493.1 APCN01002347 AAAB01008807 EAA45498.1 GL438568 EFN68767.1 ADMH02001109 ETN64084.1 DS231933 EDS27558.1 KQ435756 KOX75917.1 GAPW01003418 JAC10180.1 GEHC01000393 JAV47252.1 BABH01018411 GAMD01002746 JAA98844.1 EU035826 ABV44728.1 KZ288229 PBC31726.1 LBMM01004589 KMQ92263.1 GEZM01027546 JAV86682.1 NNAY01001573 OXU23549.1 AGBW02010081 OWR49201.1 LBMM01013914 KMQ85419.1 GGFL01005406 MBW69584.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000037510 UP000076502 UP000000311 UP000092462 UP000002320 UP000002358 UP000053097 UP000279307 UP000005203 UP000075840 UP000007062 UP000000673 UP000053105 UP000076407 UP000242457 UP000036403 UP000075882 UP000215335 UP000075902 UP000075903
UP000037510 UP000076502 UP000000311 UP000092462 UP000002320 UP000002358 UP000053097 UP000279307 UP000005203 UP000075840 UP000007062 UP000000673 UP000053105 UP000076407 UP000242457 UP000036403 UP000075882 UP000215335 UP000075902 UP000075903
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9JFI4
H9BVM5
H9JFI3
H9JFI2
B6CME8
B4X984
+ More
A4F5H5 A0A2A4J186 C9W8H0 A0A2H1VW00 A0A0X9GKP8 A0A2A4J150 A5CG73 A5CG74 A0A194PCH8 A0A2H1WEC1 H9JFI1 E7D011 A0A2H1VHU4 I6SW54 A0A1E1WV71 A5CG72 A0A0N1PHG9 A0A2H1VG38 A0A1E1W396 A0A3S2M0D3 A0A2H1VIU2 A0A212F2F5 A0A2H1VG36 A0A0N0PD42 A0A2H1VAF3 A0A194PCP7 A0A212FHN1 A0A212F2H0 A0A0L7LAZ5 A0A2H1VHM1 A0A212FIQ7 A0A0N0PDI4 A0A194PIR8 A0A2H1VG24 A0A0L7LGT4 A0A2H1WNQ2 A0A3S2LJS9 A0A2H1WSA0 H9JFE5 A0A212FHN6 A0A1E1WG09 A0A2A4JX80 A0A0L7L2A5 I6TEX4 A0A2W1BP48 Q3ZJD2 A0A194PE67 A0A212F7H7 A0A2H1X2T2 A0A2A4J0U4 A0A2W1BHD3 A0A2H1X2X6 A0A0N0PD79 A0A1Q3FSI0 A0A212F7I7 A0A154P6Q3 E1ZWT5 A0A1B0GNP3 Q9XY45 B0WW46 A0A1Q3FHD4 A0A1Q3FNV0 A0A3S2NS83 K7IRM7 A0A023EMN4 B0WLB8 Q8T639 A0A026WVM2 A0A087ZQ85 O97099 A0A182I4T2 Q7PF16 E2ACM3 W5JMT6 B0WHA0 A0A0M9A2K3 A0A182X1C1 A0A023EMU9 A0A1W7R7W0 H9JWP8 T1DGK1 A8CA02 A0A2A3EKL4 A0A0J7KPH6 A0A182KLF3 A0A1Y1MLX5 A0A232EZ15 A0A212F633 A0A0J7K549 A0A2M4CW71 A0A1S4FE82 A0A182UFJ4 A0A182V9J9
A4F5H5 A0A2A4J186 C9W8H0 A0A2H1VW00 A0A0X9GKP8 A0A2A4J150 A5CG73 A5CG74 A0A194PCH8 A0A2H1WEC1 H9JFI1 E7D011 A0A2H1VHU4 I6SW54 A0A1E1WV71 A5CG72 A0A0N1PHG9 A0A2H1VG38 A0A1E1W396 A0A3S2M0D3 A0A2H1VIU2 A0A212F2F5 A0A2H1VG36 A0A0N0PD42 A0A2H1VAF3 A0A194PCP7 A0A212FHN1 A0A212F2H0 A0A0L7LAZ5 A0A2H1VHM1 A0A212FIQ7 A0A0N0PDI4 A0A194PIR8 A0A2H1VG24 A0A0L7LGT4 A0A2H1WNQ2 A0A3S2LJS9 A0A2H1WSA0 H9JFE5 A0A212FHN6 A0A1E1WG09 A0A2A4JX80 A0A0L7L2A5 I6TEX4 A0A2W1BP48 Q3ZJD2 A0A194PE67 A0A212F7H7 A0A2H1X2T2 A0A2A4J0U4 A0A2W1BHD3 A0A2H1X2X6 A0A0N0PD79 A0A1Q3FSI0 A0A212F7I7 A0A154P6Q3 E1ZWT5 A0A1B0GNP3 Q9XY45 B0WW46 A0A1Q3FHD4 A0A1Q3FNV0 A0A3S2NS83 K7IRM7 A0A023EMN4 B0WLB8 Q8T639 A0A026WVM2 A0A087ZQ85 O97099 A0A182I4T2 Q7PF16 E2ACM3 W5JMT6 B0WHA0 A0A0M9A2K3 A0A182X1C1 A0A023EMU9 A0A1W7R7W0 H9JWP8 T1DGK1 A8CA02 A0A2A3EKL4 A0A0J7KPH6 A0A182KLF3 A0A1Y1MLX5 A0A232EZ15 A0A212F633 A0A0J7K549 A0A2M4CW71 A0A1S4FE82 A0A182UFJ4 A0A182V9J9
PDB
1EQ9
E-value=6.26002e-31,
Score=333
Ontologies
GO
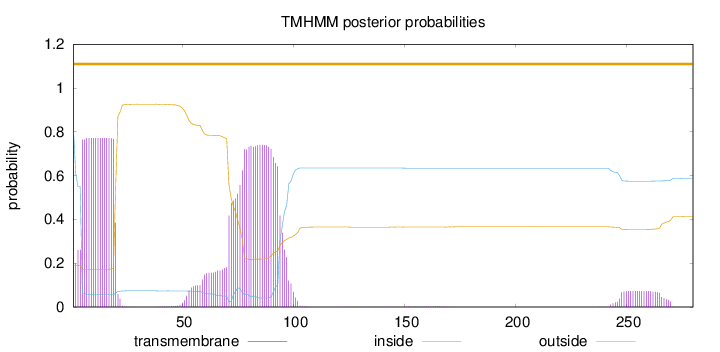
Topology
SignalP
Position: 1 - 18,
Likelihood: 0.995283

Length:
280
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
33.6976000000001
Exp number, first 60 AAs:
13.73001
Total prob of N-in:
0.80984
POSSIBLE N-term signal
sequence
outside
1 - 280
Population Genetic Test Statistics
Pi
215.123743
Theta
159.830477
Tajima's D
1.12565
CLR
1.354785
CSRT
0.687015649217539
Interpretation
Uncertain
