Gene
KWMTBOMO10710
Annotation
uncharacterized_protein_Dere_GG18014?_isoform_C_[Drosophila_erecta]
Location in the cell
PlasmaMembrane Reliability : 3.673
Sequence
CDS
ATGTGGAATCGGGAGGGGTACGAATCTGGTCGGGCCAACGTATTGGGCTGCGCCCCCCAGGCCCTTTCTATCCACTTTAGCGGTCACAATGGGCCTGTCGAGGAGCCATTCGTAGTATACGAAACTAACTGTACTTTTGAAAATGGAACGTGCAGTGACGTAGAGTACAATGTCTGGGCGCTGTCGCTACTGGTTTTTCCAGTTCTGACGCTCTTTGGTAATGTTTTGGTCATACTTAGCGTGGCAAGGGAGAGGAGTCTGCAAACAGCCACAAACTACTTCATTGTCAGTCTGGCGGTGGCCGATCTGCTTGTGGCCGTCGTGGTAATGCCGTTTGGCGTTTATTATTTGGTAAGTTGGACACGTGGTGTCAATTTTTTTTTTTTTATTGCTTAG
Protein
MWNREGYESGRANVLGCAPQALSIHFSGHNGPVEEPFVVYETNCTFENGTCSDVEYNVWALSLLVFPVLTLFGNVLVILSVARERSLQTATNYFIVSLAVADLLVAVVVMPFGVYYLVSWTRGVNFFFFIA
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
EMBL
KY092435
ATP13230.1
KP784319
AKR18180.1
AGBW02014289
OWR41932.1
+ More
CVRI01000052 CRK99666.1 CRK99667.1 AJWK01003701 AJWK01003702 AJWK01003703 AJWK01003704 KQ459606 KPI90993.1 JN859085 AFH53996.1 JRES01001467 KNC22659.1 KU710378 AOG14374.1 ABLF02038299 GGMR01002021 MBY14640.1 GFXV01004311 MBW16116.1 AAZX01002686 AAZX01002687 AAZX01005132 AAZX01005492 AAZX01005518 AAZX01010816 AAZX01011767 AAZX01011906 AAZX01013726 AAZX01025392
CVRI01000052 CRK99666.1 CRK99667.1 AJWK01003701 AJWK01003702 AJWK01003703 AJWK01003704 KQ459606 KPI90993.1 JN859085 AFH53996.1 JRES01001467 KNC22659.1 KU710378 AOG14374.1 ABLF02038299 GGMR01002021 MBY14640.1 GFXV01004311 MBW16116.1 AAZX01002686 AAZX01002687 AAZX01005132 AAZX01005492 AAZX01005518 AAZX01010816 AAZX01011767 AAZX01011906 AAZX01013726 AAZX01025392
Proteomes
Pfam
PF00001 7tm_1
ProteinModelPortal
PDB
6CM4
E-value=2.22862e-05,
Score=107
Ontologies
GO
Topology
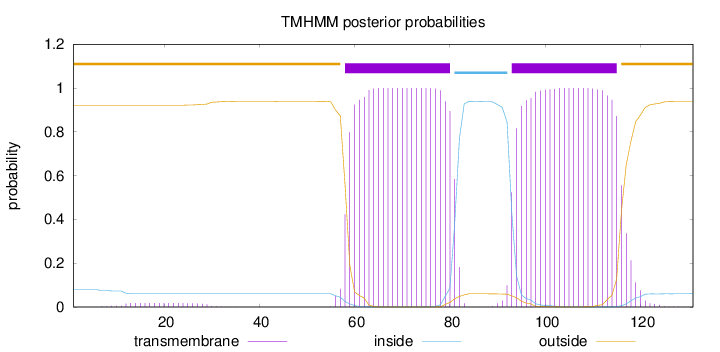
Length:
131
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
46.50351
Exp number, first 60 AAs:
2.62054
Total prob of N-in:
0.07861
outside
1 - 57
TMhelix
58 - 80
inside
81 - 92
TMhelix
93 - 115
outside
116 - 131
Population Genetic Test Statistics
Pi
44.515627
Theta
60.706113
Tajima's D
-0.789595
CLR
0.359612
CSRT
0.177741112944353
Interpretation
Uncertain
