Gene
KWMTBOMO10701
Pre Gene Modal
BGIBMGA008244
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_xanthine_dehydrogenase_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.009 Extracellular Reliability : 1.397 Nuclear Reliability : 1.294
Sequence
CDS
ATGTGTCGTGAGGCTGGGTGCGGAGCTTGCATTGTGTCTGTCGTCAAATGCCCAGGAGGACCGGTCGAAGCTGTTAATTCGTGTATGGTATCAATTACGTCATGTCAAGATTGGGAAATTACAACGATAGAAGAATTGGGGAACAGAAACAAAGGGTACAACGTTCTACAGAAGACTTTGGCTGATTATAATGGGACCCAATGTGGGTATTGCTCTCCAGCTTGGGTCATGGCGATGCACAGTCTCCTCAAAGGCAACAATGATTTAACGATGCTAGAAATAGAGAAATCCCTCAGCAGCAATGTCTGCAGATGTACTGGCTACAGACCCATATTGGACGCCTTCAAAAAATTTGCCAAAGACTGCCCTAAAGAGCATAGAATTTTAGACATAGAAGATTTGACTATCTGTAAGAGAACTGGTAAAAAATGTGATAAGAGTAATTGTAATGACGAAGACTGGTGCATAATTGAAAAAGAAGAATACAAGGCCGAGGGTATTATAGAAATACCGTTGCATGACAATAAATATTGGTACCGTGTTACGAATGTTGATGATATATTCAAAATTCTTAATCAGAAAGGATATGACTCATATATGCTGGTCTTTGGGAATACGGCTAAAGGTGCATATCCAATCTTCGAATATCCGAAAGTATTGATTGATATTAGCGCTGTAGCCCAATTAAAGATTTGCCTTTATGACCAGAATTTAGTAGTAGGTGTTGGATTGACGCTTACCGAATTATTGAATACTTTCGAGACAGCTTCAAAGGAAGAATACTTCAAGTACCTGAGGAAGCTTTACGAACATTTGTTATTGGTCGCCCACATACCCGTCAGAAATGTATGTAAATCGATACTAAGTTGTATGTTGTATCAAAAACGAAATCCATACAATAAGTAA
Protein
MCREAGCGACIVSVVKCPGGPVEAVNSCMVSITSCQDWEITTIEELGNRNKGYNVLQKTLADYNGTQCGYCSPAWVMAMHSLLKGNNDLTMLEIEKSLSSNVCRCTGYRPILDAFKKFAKDCPKEHRILDIEDLTICKRTGKKCDKSNCNDEDWCIIEKEEYKAEGIIEIPLHDNKYWYRVTNVDDIFKILNQKGYDSYMLVFGNTAKGAYPIFEYPKVLIDISAVAQLKICLYDQNLVVGVGLTLTELLNTFETASKEEYFKYLRKLYEHLLLVAHIPVRNVCKSILSCMLYQKRNPYNK
Summary
Cofactor
FAD
Mo-molybdopterin
[2Fe-2S] cluster
Mo-molybdopterin
[2Fe-2S] cluster
Uniprot
H9JFE7
A0A2W1BGZ9
A0A0N0PD45
A0A2H1VZA4
A0A3S2PBH0
A0A2W1BJJ3
+ More
A0A0L7L4E9 A0A2A4IRV9 A0A2A4ISI6 A0A2A4ITB4 A0A0F7QIE4 A0A2H1V6B6 A0A194PE80 A0A2H1V2I4 A0A3S2M0B9 A0A0N0PDI7 A0A2W1BIK1 A0A2A4JU78 A0A194PCQ5 A0A212EVU1 A0A2H1VA65 A0A194PDB3 A0A194PIT3 A0A2A4JUF7 A0A0N1IAK3 A0A3S2NDD1 A0A1X9ZM11 A0A0L7L438 A0A2W1BIE3 A0A2W1BHI1 A0A212FI14 A0A2A4ITG3 H9JFH1 A0A0L7L4A2 A0A1X9ZM28 A0A2H1VDX2 A0A194QBY5 A0A0L7L3S0 A0A0N0PF58 A0A194QUG5 A0A212EZ25 A0A2A4IZZ7 A0A2H1WKN9 A0A0H4TD12 A0A2H1VSX5 A0A2H1VA49 A0A194PIN2 A0A076FRM9 A0A0F7QIE0 A0A1L8D6P5 Q4VGM3 A0A0F7QEA3 A0A3S2P6B3 A0A2H1WDE7 A0A2H1VCW8 A0A1L8D6Q4 A0A2A4KA50 A0A1X9ZM23 A0A212FKB8 A0A212F791 A0A2H1VH28 A0A194PDZ5 A0A194PCA1 S5FPI8 A0A194QM53 A0A2A4JXM9 A0A1U9X1S0 A0A0F7QJV6 A0A2A4JYR8 A0A0F7QEA1 A0A2H1V7P8 A0A0H4TY60 H9JFZ8 H9JFZ2 A8TUB4 A0A194QMP0 A0A076FX80 A0A1B6G9B1 A0A0A9YA60 A0A1X9ZLZ6 A0A0N1I7K2 A0A3S2NMY0 A0A1B6E3D0 A0A146L5G6 A0A0A9YHM8 A0A146MB53 A0A154PJ92 A0A336MA50 T1HWZ8 H9JFZ3 A0A2A4JX39 A0A076FRA1 A0A1B6I539 A8TUC0 K7ITY0 A0A1Y9H2G7 A0A023F0C3 W5JCY6
A0A0L7L4E9 A0A2A4IRV9 A0A2A4ISI6 A0A2A4ITB4 A0A0F7QIE4 A0A2H1V6B6 A0A194PE80 A0A2H1V2I4 A0A3S2M0B9 A0A0N0PDI7 A0A2W1BIK1 A0A2A4JU78 A0A194PCQ5 A0A212EVU1 A0A2H1VA65 A0A194PDB3 A0A194PIT3 A0A2A4JUF7 A0A0N1IAK3 A0A3S2NDD1 A0A1X9ZM11 A0A0L7L438 A0A2W1BIE3 A0A2W1BHI1 A0A212FI14 A0A2A4ITG3 H9JFH1 A0A0L7L4A2 A0A1X9ZM28 A0A2H1VDX2 A0A194QBY5 A0A0L7L3S0 A0A0N0PF58 A0A194QUG5 A0A212EZ25 A0A2A4IZZ7 A0A2H1WKN9 A0A0H4TD12 A0A2H1VSX5 A0A2H1VA49 A0A194PIN2 A0A076FRM9 A0A0F7QIE0 A0A1L8D6P5 Q4VGM3 A0A0F7QEA3 A0A3S2P6B3 A0A2H1WDE7 A0A2H1VCW8 A0A1L8D6Q4 A0A2A4KA50 A0A1X9ZM23 A0A212FKB8 A0A212F791 A0A2H1VH28 A0A194PDZ5 A0A194PCA1 S5FPI8 A0A194QM53 A0A2A4JXM9 A0A1U9X1S0 A0A0F7QJV6 A0A2A4JYR8 A0A0F7QEA1 A0A2H1V7P8 A0A0H4TY60 H9JFZ8 H9JFZ2 A8TUB4 A0A194QMP0 A0A076FX80 A0A1B6G9B1 A0A0A9YA60 A0A1X9ZLZ6 A0A0N1I7K2 A0A3S2NMY0 A0A1B6E3D0 A0A146L5G6 A0A0A9YHM8 A0A146MB53 A0A154PJ92 A0A336MA50 T1HWZ8 H9JFZ3 A0A2A4JX39 A0A076FRA1 A0A1B6I539 A8TUC0 K7ITY0 A0A1Y9H2G7 A0A023F0C3 W5JCY6
Pubmed
EMBL
BABH01034979
BABH01034980
BABH01034981
KZ150135
PZC72995.1
KQ460326
+ More
KPJ15694.1 ODYU01005369 SOQ46133.1 RSAL01000123 RVE46648.1 PZC72996.1 JTDY01003022 KOB70290.1 NWSH01007964 PCG62737.1 PCG62735.1 PCG62736.1 LC017759 BAR64773.1 ODYU01000882 SOQ36316.1 KQ459606 KPI90999.1 ODYU01000376 SOQ35055.1 RSAL01000088 RVE48203.1 KPJ15692.1 KZ150165 PZC72656.1 NWSH01000648 PCG75033.1 KPI90997.1 AGBW02012116 OWR45615.1 ODYU01001481 SOQ37735.1 KPI90998.1 KPI91000.1 PCG75032.1 KPJ15693.1 RVE48202.1 KY595781 ARS46835.1 JTDY01003130 KOB70096.1 PZC72997.1 PZC72657.1 AGBW02008446 OWR53382.1 NWSH01007172 PCG63055.1 BABH01034958 BABH01034959 BABH01034960 BABH01034961 BABH01034962 BABH01034963 BABH01034964 BABH01034965 JTDY01003017 KOB70302.1 KY595780 ARS46834.1 ODYU01002030 SOQ39029.1 KQ459193 KPJ03063.1 JTDY01003126 KOB70107.1 KQ459674 KPJ20482.1 KQ461154 KPJ08620.1 AGBW02011403 OWR46707.1 NWSH01004179 PCG65395.1 ODYU01009314 SOQ53643.1 KP899546 AKQ06145.1 ODYU01004253 SOQ43930.1 ODYU01001472 SOQ37709.1 KPI90950.1 KF960796 AII21998.1 LC017754 BAR64768.1 GEYN01000003 JAV02126.1 AY947538 AAY22446.1 LC017757 BAR64771.1 RSAL01000295 RVE42846.1 ODYU01007852 SOQ50986.1 ODYU01001878 SOQ38698.1 GEYN01000001 JAV02128.1 NWSH01000022 PCG80663.1 KY595779 ARS46833.1 AGBW02008082 OWR54176.1 AGBW02009901 OWR49607.1 ODYU01002537 SOQ40155.1 KPI90909.1 KPI90901.1 KC952900 AGQ43599.1 KQ461196 KPJ06623.1 NWSH01000413 PCG76559.1 KY643683 AQY62686.1 LC017753 BAR64767.1 PCG76560.1 LC017752 BAR64766.1 ODYU01001109 SOQ36868.1 KP899547 AKQ06146.1 BABH01001678 BABH01001679 BABH01001657 BABH01001658 BABH01001659 BABH01001660 EU073423 ABW71271.1 KPJ06614.1 KF960794 AII21996.1 GECZ01010778 JAS58991.1 GBHO01014555 GBHO01014552 JAG29049.1 JAG29052.1 KY595778 ARS46832.1 KQ460556 KPJ13979.1 RSAL01000229 RVE43893.1 GEDC01004862 JAS32436.1 GDHC01015520 JAQ03109.1 GBHO01014557 GBHO01014554 GBRD01002756 JAG29047.1 JAG29050.1 JAG63065.1 GDHC01013650 GDHC01001960 JAQ04979.1 JAQ16669.1 KQ434912 KZC11368.1 UFQS01000745 UFQT01000745 SSX06553.1 SSX26900.1 ACPB03000354 ACPB03000355 BABH01001661 PCG76561.1 KF960795 AII21997.1 GECU01025650 JAS82056.1 EU073424 ABW71272.1 GBBI01004413 JAC14299.1 ADMH02001746 ETN61218.1
KPJ15694.1 ODYU01005369 SOQ46133.1 RSAL01000123 RVE46648.1 PZC72996.1 JTDY01003022 KOB70290.1 NWSH01007964 PCG62737.1 PCG62735.1 PCG62736.1 LC017759 BAR64773.1 ODYU01000882 SOQ36316.1 KQ459606 KPI90999.1 ODYU01000376 SOQ35055.1 RSAL01000088 RVE48203.1 KPJ15692.1 KZ150165 PZC72656.1 NWSH01000648 PCG75033.1 KPI90997.1 AGBW02012116 OWR45615.1 ODYU01001481 SOQ37735.1 KPI90998.1 KPI91000.1 PCG75032.1 KPJ15693.1 RVE48202.1 KY595781 ARS46835.1 JTDY01003130 KOB70096.1 PZC72997.1 PZC72657.1 AGBW02008446 OWR53382.1 NWSH01007172 PCG63055.1 BABH01034958 BABH01034959 BABH01034960 BABH01034961 BABH01034962 BABH01034963 BABH01034964 BABH01034965 JTDY01003017 KOB70302.1 KY595780 ARS46834.1 ODYU01002030 SOQ39029.1 KQ459193 KPJ03063.1 JTDY01003126 KOB70107.1 KQ459674 KPJ20482.1 KQ461154 KPJ08620.1 AGBW02011403 OWR46707.1 NWSH01004179 PCG65395.1 ODYU01009314 SOQ53643.1 KP899546 AKQ06145.1 ODYU01004253 SOQ43930.1 ODYU01001472 SOQ37709.1 KPI90950.1 KF960796 AII21998.1 LC017754 BAR64768.1 GEYN01000003 JAV02126.1 AY947538 AAY22446.1 LC017757 BAR64771.1 RSAL01000295 RVE42846.1 ODYU01007852 SOQ50986.1 ODYU01001878 SOQ38698.1 GEYN01000001 JAV02128.1 NWSH01000022 PCG80663.1 KY595779 ARS46833.1 AGBW02008082 OWR54176.1 AGBW02009901 OWR49607.1 ODYU01002537 SOQ40155.1 KPI90909.1 KPI90901.1 KC952900 AGQ43599.1 KQ461196 KPJ06623.1 NWSH01000413 PCG76559.1 KY643683 AQY62686.1 LC017753 BAR64767.1 PCG76560.1 LC017752 BAR64766.1 ODYU01001109 SOQ36868.1 KP899547 AKQ06146.1 BABH01001678 BABH01001679 BABH01001657 BABH01001658 BABH01001659 BABH01001660 EU073423 ABW71271.1 KPJ06614.1 KF960794 AII21996.1 GECZ01010778 JAS58991.1 GBHO01014555 GBHO01014552 JAG29049.1 JAG29052.1 KY595778 ARS46832.1 KQ460556 KPJ13979.1 RSAL01000229 RVE43893.1 GEDC01004862 JAS32436.1 GDHC01015520 JAQ03109.1 GBHO01014557 GBHO01014554 GBRD01002756 JAG29047.1 JAG29050.1 JAG63065.1 GDHC01013650 GDHC01001960 JAQ04979.1 JAQ16669.1 KQ434912 KZC11368.1 UFQS01000745 UFQT01000745 SSX06553.1 SSX26900.1 ACPB03000354 ACPB03000355 BABH01001661 PCG76561.1 KF960795 AII21997.1 GECU01025650 JAS82056.1 EU073424 ABW71272.1 GBBI01004413 JAC14299.1 ADMH02001746 ETN61218.1
Proteomes
Pfam
Interpro
IPR002888
2Fe-2S-bd
+ More
IPR016208 Ald_Oxase/xanthine_DH
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR036318 FAD-bd_PCMH-like_sf
IPR036884 2Fe-2S-bd_dom_sf
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR005107 CO_DH_flav_C
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR002346 Mopterin_DH_FAD-bd
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR012675 Beta-grasp_dom_sf
IPR016169 FAD-bd_PCMH_sub2
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR002159 CD36_fam
IPR016208 Ald_Oxase/xanthine_DH
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR036318 FAD-bd_PCMH-like_sf
IPR036884 2Fe-2S-bd_dom_sf
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR005107 CO_DH_flav_C
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR002346 Mopterin_DH_FAD-bd
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR012675 Beta-grasp_dom_sf
IPR016169 FAD-bd_PCMH_sub2
IPR027417 P-loop_NTPase
IPR002464 DNA/RNA_helicase_DEAH_CS
IPR014001 Helicase_ATP-bd
IPR001650 Helicase_C
IPR002159 CD36_fam
SUPFAM
Gene 3D
ProteinModelPortal
H9JFE7
A0A2W1BGZ9
A0A0N0PD45
A0A2H1VZA4
A0A3S2PBH0
A0A2W1BJJ3
+ More
A0A0L7L4E9 A0A2A4IRV9 A0A2A4ISI6 A0A2A4ITB4 A0A0F7QIE4 A0A2H1V6B6 A0A194PE80 A0A2H1V2I4 A0A3S2M0B9 A0A0N0PDI7 A0A2W1BIK1 A0A2A4JU78 A0A194PCQ5 A0A212EVU1 A0A2H1VA65 A0A194PDB3 A0A194PIT3 A0A2A4JUF7 A0A0N1IAK3 A0A3S2NDD1 A0A1X9ZM11 A0A0L7L438 A0A2W1BIE3 A0A2W1BHI1 A0A212FI14 A0A2A4ITG3 H9JFH1 A0A0L7L4A2 A0A1X9ZM28 A0A2H1VDX2 A0A194QBY5 A0A0L7L3S0 A0A0N0PF58 A0A194QUG5 A0A212EZ25 A0A2A4IZZ7 A0A2H1WKN9 A0A0H4TD12 A0A2H1VSX5 A0A2H1VA49 A0A194PIN2 A0A076FRM9 A0A0F7QIE0 A0A1L8D6P5 Q4VGM3 A0A0F7QEA3 A0A3S2P6B3 A0A2H1WDE7 A0A2H1VCW8 A0A1L8D6Q4 A0A2A4KA50 A0A1X9ZM23 A0A212FKB8 A0A212F791 A0A2H1VH28 A0A194PDZ5 A0A194PCA1 S5FPI8 A0A194QM53 A0A2A4JXM9 A0A1U9X1S0 A0A0F7QJV6 A0A2A4JYR8 A0A0F7QEA1 A0A2H1V7P8 A0A0H4TY60 H9JFZ8 H9JFZ2 A8TUB4 A0A194QMP0 A0A076FX80 A0A1B6G9B1 A0A0A9YA60 A0A1X9ZLZ6 A0A0N1I7K2 A0A3S2NMY0 A0A1B6E3D0 A0A146L5G6 A0A0A9YHM8 A0A146MB53 A0A154PJ92 A0A336MA50 T1HWZ8 H9JFZ3 A0A2A4JX39 A0A076FRA1 A0A1B6I539 A8TUC0 K7ITY0 A0A1Y9H2G7 A0A023F0C3 W5JCY6
A0A0L7L4E9 A0A2A4IRV9 A0A2A4ISI6 A0A2A4ITB4 A0A0F7QIE4 A0A2H1V6B6 A0A194PE80 A0A2H1V2I4 A0A3S2M0B9 A0A0N0PDI7 A0A2W1BIK1 A0A2A4JU78 A0A194PCQ5 A0A212EVU1 A0A2H1VA65 A0A194PDB3 A0A194PIT3 A0A2A4JUF7 A0A0N1IAK3 A0A3S2NDD1 A0A1X9ZM11 A0A0L7L438 A0A2W1BIE3 A0A2W1BHI1 A0A212FI14 A0A2A4ITG3 H9JFH1 A0A0L7L4A2 A0A1X9ZM28 A0A2H1VDX2 A0A194QBY5 A0A0L7L3S0 A0A0N0PF58 A0A194QUG5 A0A212EZ25 A0A2A4IZZ7 A0A2H1WKN9 A0A0H4TD12 A0A2H1VSX5 A0A2H1VA49 A0A194PIN2 A0A076FRM9 A0A0F7QIE0 A0A1L8D6P5 Q4VGM3 A0A0F7QEA3 A0A3S2P6B3 A0A2H1WDE7 A0A2H1VCW8 A0A1L8D6Q4 A0A2A4KA50 A0A1X9ZM23 A0A212FKB8 A0A212F791 A0A2H1VH28 A0A194PDZ5 A0A194PCA1 S5FPI8 A0A194QM53 A0A2A4JXM9 A0A1U9X1S0 A0A0F7QJV6 A0A2A4JYR8 A0A0F7QEA1 A0A2H1V7P8 A0A0H4TY60 H9JFZ8 H9JFZ2 A8TUB4 A0A194QMP0 A0A076FX80 A0A1B6G9B1 A0A0A9YA60 A0A1X9ZLZ6 A0A0N1I7K2 A0A3S2NMY0 A0A1B6E3D0 A0A146L5G6 A0A0A9YHM8 A0A146MB53 A0A154PJ92 A0A336MA50 T1HWZ8 H9JFZ3 A0A2A4JX39 A0A076FRA1 A0A1B6I539 A8TUC0 K7ITY0 A0A1Y9H2G7 A0A023F0C3 W5JCY6
PDB
4YTZ
E-value=1.05531e-21,
Score=254
Ontologies
PATHWAY
GO
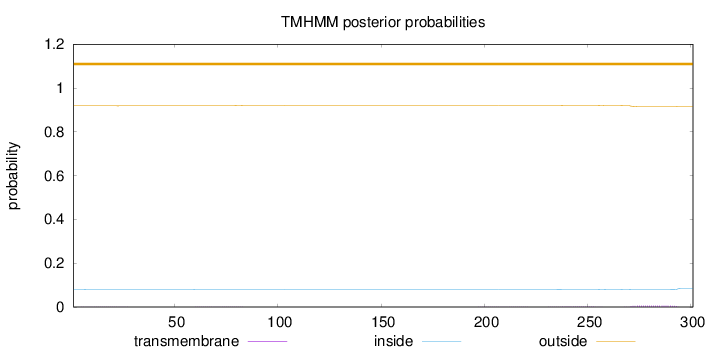
Topology
Length:
301
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20257
Exp number, first 60 AAs:
0.01287
Total prob of N-in:
0.08122
outside
1 - 301
Population Genetic Test Statistics
Pi
35.408983
Theta
53.474056
Tajima's D
0.019145
CLR
0.647227
CSRT
0.373531323433828
Interpretation
Uncertain
