Gene
KWMTBOMO10699
Pre Gene Modal
BGIBMGA008244
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_xanthine_dehydrogenase_1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.244 Mitochondrial Reliability : 1.102
Sequence
CDS
ATGTCTGATACTCTTAAAAAAAACGCTGAGTATACCAAAAGGCGTGCCGCTGTCACGAAATTCAATGCTGAAAACAGATGGAAGAAACGTGGCCTTAGGTGCTCCTTCTCTAGGTGGACGCCAGCAGGAGGTCAGTACTTGGACACAAACATGGCAGTCTACCACGAAGATGGTACTGTTGCCATTTGCCACGGTGGCATAGAAATGGGCCAAGGGCTGAACACAAAAGCGGCCCAAATCTGTGCGTATTACTTAAAGATACCCTTAGAGAAAATTCAGGTTAAGGCCAACAGCACGGTTTTGACACCTAACGGCTTTATATCCGGTGGAAGTCAGACGTCACTGAATATAGGCGTAGCTGTTACGAGGTGTTGTGAGGAACTGTTACGAAGATTGCAGCCAATAAGATTGCAGATGAACAACCCGACTTGGGAGGAATTAGTGAAGAAAGCTTTTGATTCTAATGTCGATTTGCAAGTTCATGGTTTCGTTAGCGCTATCGATGCACAGATCTATAATATTTTCGGAGTTTCCTTGGCAGAAGTTGAGGTGGACGTTTTAACTGGAGAGTCGGAGATATTAAGAGTCGATATCCTACAAGACGTCGGGAAAAGCGTCAGCCCTGAAATCGATATCGGACAAGTTGAAGGGGCATTCATCATGGGTCTTGGGTACTGGACATGTGAGAAGTTGGTATATGAACCTAATACTGGGGAATTACTCACTGACCGCACTTGGAACTATAGCGTGCCCTTAGCAAGAGATATCCCGCAAGATCTCAGAGTATACTTCAGGAAGAACTCTTACAGTACTGAACTTTTTCTTGGAGCCAAAGGTACGCTTGTATTGCTATTGTCAAGAATGAATGAATGA
Protein
MSDTLKKNAEYTKRRAAVTKFNAENRWKKRGLRCSFSRWTPAGGQYLDTNMAVYHEDGTVAICHGGIEMGQGLNTKAAQICAYYLKIPLEKIQVKANSTVLTPNGFISGGSQTSLNIGVAVTRCCEELLRRLQPIRLQMNNPTWEELVKKAFDSNVDLQVHGFVSAIDAQIYNIFGVSLAEVEVDVLTGESEILRVDILQDVGKSVSPEIDIGQVEGAFIMGLGYWTCEKLVYEPNTGELLTDRTWNYSVPLARDIPQDLRVYFRKNSYSTELFLGAKGTLVLLLSRMNE
Summary
Cofactor
FAD
Mo-molybdopterin
[2Fe-2S] cluster
Mo-molybdopterin
[2Fe-2S] cluster
Uniprot
H9JFE7
A0A2W1BG03
A0A194PIT3
A0A2H1VNQ0
A0A2W1BIK1
A0A0N0PD45
+ More
A0A2A4JU78 A0A2A4JUF7 A0A0F7QJW1 A0A3S2PBH0 A0A2W1BHI1 A0A194PDB3 A0A1X9ZM11 A0A0F7QEC7 A0A0L7L438 A0A0N1IAK3 A0A2W1BH31 A0A2H1VEC0 A0A2H1V6B6 A0A194PE80 A0A2A4ITB4 A0A2W1BJJ3 A0A0L7L3S0 A0A2H1VA65 A0A194PCQ5 A0A0N0PDI7 A0A3S2NDD1 A0A2A4ISI6 A0A212FI14 A0A2W1BIE3 A0A2H1V2I4 A0A3S2NTK8 A0A0L7L4E9 A0A2A4IRV9 H9JFH1 A0A1X9ZM28 A0A212EZ25 A0A1B6E8K7 A0A0F7QEA3 A0A194QCB6 A0A2H1V098 A0A2H1VUX4 A0A0N0PF58 A0A0F7QHK5 H9JFX1 A0A2A4ITG3 A0A2J7PXR7 A0A0L7L4A2 A0A2H1VH28 A0A2H1V0B2 A0A2H1VCW8 A0A076FX80 A0A2H1VSX5 A0A2A4JXM9 A0A2A4JB30 H9JFZ3 A0A1B6C5G4 H9JFZ2 A0A1B6I7V1 A0A1B6CXI3 A0A2A4JYR8 A8TUC0 A0A2H1V7P8 A0A076FRA1 H9JFX4 A0A067R2R7 A0A2A4JX39 A0A194QBY5 A0A194PIN2 A0A2C9GUL3 A0A2M4CMZ5 A0A0F7QEA1 A0A1Y9H2G2 A0A1Y9J0A2 W5JEG5 A0A182QVB6 A0A182R8R0 A0A212F791 A0A194QUG5 A0A2A4JY06 A0A182U9S4 A0A1L8DXJ0 A0A0V0G7W2 A0A1B6FB46 A0A1B6JH49 A0A240PL19 A0A182LLK4 A0A194PDZ5 A0A1L8DXS0 A0A3S2NMY0 A0A151X3M2 A0A182JVC0 A0A1L8DXU8 A0A0A9Y5G8 A0A195E3I3 A0A1B6JA43 A0A2M4BAX5 A0A2M4BAZ2 A0A182YKY2 A0A1Y9HAI9
A0A2A4JU78 A0A2A4JUF7 A0A0F7QJW1 A0A3S2PBH0 A0A2W1BHI1 A0A194PDB3 A0A1X9ZM11 A0A0F7QEC7 A0A0L7L438 A0A0N1IAK3 A0A2W1BH31 A0A2H1VEC0 A0A2H1V6B6 A0A194PE80 A0A2A4ITB4 A0A2W1BJJ3 A0A0L7L3S0 A0A2H1VA65 A0A194PCQ5 A0A0N0PDI7 A0A3S2NDD1 A0A2A4ISI6 A0A212FI14 A0A2W1BIE3 A0A2H1V2I4 A0A3S2NTK8 A0A0L7L4E9 A0A2A4IRV9 H9JFH1 A0A1X9ZM28 A0A212EZ25 A0A1B6E8K7 A0A0F7QEA3 A0A194QCB6 A0A2H1V098 A0A2H1VUX4 A0A0N0PF58 A0A0F7QHK5 H9JFX1 A0A2A4ITG3 A0A2J7PXR7 A0A0L7L4A2 A0A2H1VH28 A0A2H1V0B2 A0A2H1VCW8 A0A076FX80 A0A2H1VSX5 A0A2A4JXM9 A0A2A4JB30 H9JFZ3 A0A1B6C5G4 H9JFZ2 A0A1B6I7V1 A0A1B6CXI3 A0A2A4JYR8 A8TUC0 A0A2H1V7P8 A0A076FRA1 H9JFX4 A0A067R2R7 A0A2A4JX39 A0A194QBY5 A0A194PIN2 A0A2C9GUL3 A0A2M4CMZ5 A0A0F7QEA1 A0A1Y9H2G2 A0A1Y9J0A2 W5JEG5 A0A182QVB6 A0A182R8R0 A0A212F791 A0A194QUG5 A0A2A4JY06 A0A182U9S4 A0A1L8DXJ0 A0A0V0G7W2 A0A1B6FB46 A0A1B6JH49 A0A240PL19 A0A182LLK4 A0A194PDZ5 A0A1L8DXS0 A0A3S2NMY0 A0A151X3M2 A0A182JVC0 A0A1L8DXU8 A0A0A9Y5G8 A0A195E3I3 A0A1B6JA43 A0A2M4BAX5 A0A2M4BAZ2 A0A182YKY2 A0A1Y9HAI9
Pubmed
EMBL
BABH01034979
BABH01034980
BABH01034981
KZ150165
PZC72655.1
KQ459606
+ More
KPI91000.1 ODYU01003306 SOQ41884.1 PZC72656.1 KQ460326 KPJ15694.1 NWSH01000648 PCG75033.1 PCG75032.1 LC017758 BAR64772.1 RSAL01000123 RVE46648.1 PZC72657.1 KPI90998.1 KY595781 ARS46835.1 LC017756 BAR64770.1 JTDY01003130 KOB70096.1 KPJ15693.1 KZ150135 PZC72994.1 ODYU01002094 SOQ39169.1 ODYU01000882 SOQ36316.1 KPI90999.1 NWSH01007964 PCG62736.1 PZC72996.1 JTDY01003126 KOB70107.1 ODYU01001481 SOQ37735.1 KPI90997.1 KPJ15692.1 RSAL01000088 RVE48202.1 PCG62735.1 AGBW02008446 OWR53382.1 PZC72997.1 ODYU01000376 SOQ35055.1 RVE48204.1 JTDY01003022 KOB70290.1 PCG62737.1 BABH01034958 BABH01034959 BABH01034960 BABH01034961 BABH01034962 BABH01034963 BABH01034964 BABH01034965 KY595780 ARS46834.1 AGBW02011403 OWR46707.1 GEDC01003026 JAS34272.1 LC017757 BAR64771.1 KQ459193 KPJ03064.1 ODYU01000094 SOQ34283.1 ODYU01004588 SOQ44620.1 KQ459674 KPJ20482.1 LC017755 BAR64769.1 BABH01001558 NWSH01007172 PCG63055.1 NEVH01020852 PNF21138.1 JTDY01003017 KOB70302.1 ODYU01002537 SOQ40155.1 ODYU01000095 SOQ34285.1 ODYU01001878 SOQ38698.1 KF960794 AII21996.1 ODYU01004253 SOQ43930.1 NWSH01000413 PCG76559.1 NWSH01002143 PCG69059.1 BABH01001661 GEDC01028540 JAS08758.1 BABH01001657 BABH01001658 BABH01001659 BABH01001660 GECU01024721 JAS82985.1 GEDC01019096 JAS18202.1 PCG76560.1 EU073424 ABW71272.1 ODYU01001109 SOQ36868.1 KF960795 AII21997.1 BABH01001568 BABH01001569 BABH01001570 BABH01001571 BABH01001572 BABH01001573 KK852750 KDR17215.1 PCG76561.1 KPJ03063.1 KPI90950.1 AXCM01000856 GGFL01002475 MBW66653.1 LC017752 BAR64766.1 ADMH02001746 ETN61219.1 AXCN02000553 AGBW02009901 OWR49607.1 KQ461154 KPJ08620.1 PCG76558.1 GFDF01002904 JAV11180.1 GECL01002669 JAP03455.1 GECZ01022332 JAS47437.1 GECU01009219 JAS98487.1 KPI90909.1 GFDF01002843 JAV11241.1 RSAL01000229 RVE43893.1 KQ982557 KYQ55015.1 GFDF01002797 JAV11287.1 GBHO01015297 JAG28307.1 KQ979701 KYN19698.1 GECU01011660 JAS96046.1 GGFJ01001064 MBW50205.1 GGFJ01001065 MBW50206.1 AXCN02000554
KPI91000.1 ODYU01003306 SOQ41884.1 PZC72656.1 KQ460326 KPJ15694.1 NWSH01000648 PCG75033.1 PCG75032.1 LC017758 BAR64772.1 RSAL01000123 RVE46648.1 PZC72657.1 KPI90998.1 KY595781 ARS46835.1 LC017756 BAR64770.1 JTDY01003130 KOB70096.1 KPJ15693.1 KZ150135 PZC72994.1 ODYU01002094 SOQ39169.1 ODYU01000882 SOQ36316.1 KPI90999.1 NWSH01007964 PCG62736.1 PZC72996.1 JTDY01003126 KOB70107.1 ODYU01001481 SOQ37735.1 KPI90997.1 KPJ15692.1 RSAL01000088 RVE48202.1 PCG62735.1 AGBW02008446 OWR53382.1 PZC72997.1 ODYU01000376 SOQ35055.1 RVE48204.1 JTDY01003022 KOB70290.1 PCG62737.1 BABH01034958 BABH01034959 BABH01034960 BABH01034961 BABH01034962 BABH01034963 BABH01034964 BABH01034965 KY595780 ARS46834.1 AGBW02011403 OWR46707.1 GEDC01003026 JAS34272.1 LC017757 BAR64771.1 KQ459193 KPJ03064.1 ODYU01000094 SOQ34283.1 ODYU01004588 SOQ44620.1 KQ459674 KPJ20482.1 LC017755 BAR64769.1 BABH01001558 NWSH01007172 PCG63055.1 NEVH01020852 PNF21138.1 JTDY01003017 KOB70302.1 ODYU01002537 SOQ40155.1 ODYU01000095 SOQ34285.1 ODYU01001878 SOQ38698.1 KF960794 AII21996.1 ODYU01004253 SOQ43930.1 NWSH01000413 PCG76559.1 NWSH01002143 PCG69059.1 BABH01001661 GEDC01028540 JAS08758.1 BABH01001657 BABH01001658 BABH01001659 BABH01001660 GECU01024721 JAS82985.1 GEDC01019096 JAS18202.1 PCG76560.1 EU073424 ABW71272.1 ODYU01001109 SOQ36868.1 KF960795 AII21997.1 BABH01001568 BABH01001569 BABH01001570 BABH01001571 BABH01001572 BABH01001573 KK852750 KDR17215.1 PCG76561.1 KPJ03063.1 KPI90950.1 AXCM01000856 GGFL01002475 MBW66653.1 LC017752 BAR64766.1 ADMH02001746 ETN61219.1 AXCN02000553 AGBW02009901 OWR49607.1 KQ461154 KPJ08620.1 PCG76558.1 GFDF01002904 JAV11180.1 GECL01002669 JAP03455.1 GECZ01022332 JAS47437.1 GECU01009219 JAS98487.1 KPI90909.1 GFDF01002843 JAV11241.1 RSAL01000229 RVE43893.1 KQ982557 KYQ55015.1 GFDF01002797 JAV11287.1 GBHO01015297 JAG28307.1 KQ979701 KYN19698.1 GECU01011660 JAS96046.1 GGFJ01001064 MBW50205.1 GGFJ01001065 MBW50206.1 AXCN02000554
Proteomes
Pfam
Interpro
IPR002888
2Fe-2S-bd
+ More
IPR016208 Ald_Oxase/xanthine_DH
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR036318 FAD-bd_PCMH-like_sf
IPR036884 2Fe-2S-bd_dom_sf
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR005107 CO_DH_flav_C
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR002346 Mopterin_DH_FAD-bd
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR016169 FAD-bd_PCMH_sub2
IPR012675 Beta-grasp_dom_sf
IPR002159 CD36_fam
IPR016208 Ald_Oxase/xanthine_DH
IPR000674 Ald_Oxase/Xan_DH_a/b
IPR036318 FAD-bd_PCMH-like_sf
IPR036884 2Fe-2S-bd_dom_sf
IPR008274 AldOxase/xan_DH_Mopterin-bd
IPR005107 CO_DH_flav_C
IPR001041 2Fe-2S_ferredoxin-type
IPR036010 2Fe-2S_ferredoxin-like_sf
IPR036683 CO_DH_flav_C_dom_sf
IPR006058 2Fe2S_fd_BS
IPR016166 FAD-bd_PCMH
IPR037165 AldOxase/xan_DH_Mopterin-bd_sf
IPR002346 Mopterin_DH_FAD-bd
IPR036856 Ald_Oxase/Xan_DH_a/b_sf
IPR016169 FAD-bd_PCMH_sub2
IPR012675 Beta-grasp_dom_sf
IPR002159 CD36_fam
SUPFAM
Gene 3D
CDD
ProteinModelPortal
H9JFE7
A0A2W1BG03
A0A194PIT3
A0A2H1VNQ0
A0A2W1BIK1
A0A0N0PD45
+ More
A0A2A4JU78 A0A2A4JUF7 A0A0F7QJW1 A0A3S2PBH0 A0A2W1BHI1 A0A194PDB3 A0A1X9ZM11 A0A0F7QEC7 A0A0L7L438 A0A0N1IAK3 A0A2W1BH31 A0A2H1VEC0 A0A2H1V6B6 A0A194PE80 A0A2A4ITB4 A0A2W1BJJ3 A0A0L7L3S0 A0A2H1VA65 A0A194PCQ5 A0A0N0PDI7 A0A3S2NDD1 A0A2A4ISI6 A0A212FI14 A0A2W1BIE3 A0A2H1V2I4 A0A3S2NTK8 A0A0L7L4E9 A0A2A4IRV9 H9JFH1 A0A1X9ZM28 A0A212EZ25 A0A1B6E8K7 A0A0F7QEA3 A0A194QCB6 A0A2H1V098 A0A2H1VUX4 A0A0N0PF58 A0A0F7QHK5 H9JFX1 A0A2A4ITG3 A0A2J7PXR7 A0A0L7L4A2 A0A2H1VH28 A0A2H1V0B2 A0A2H1VCW8 A0A076FX80 A0A2H1VSX5 A0A2A4JXM9 A0A2A4JB30 H9JFZ3 A0A1B6C5G4 H9JFZ2 A0A1B6I7V1 A0A1B6CXI3 A0A2A4JYR8 A8TUC0 A0A2H1V7P8 A0A076FRA1 H9JFX4 A0A067R2R7 A0A2A4JX39 A0A194QBY5 A0A194PIN2 A0A2C9GUL3 A0A2M4CMZ5 A0A0F7QEA1 A0A1Y9H2G2 A0A1Y9J0A2 W5JEG5 A0A182QVB6 A0A182R8R0 A0A212F791 A0A194QUG5 A0A2A4JY06 A0A182U9S4 A0A1L8DXJ0 A0A0V0G7W2 A0A1B6FB46 A0A1B6JH49 A0A240PL19 A0A182LLK4 A0A194PDZ5 A0A1L8DXS0 A0A3S2NMY0 A0A151X3M2 A0A182JVC0 A0A1L8DXU8 A0A0A9Y5G8 A0A195E3I3 A0A1B6JA43 A0A2M4BAX5 A0A2M4BAZ2 A0A182YKY2 A0A1Y9HAI9
A0A2A4JU78 A0A2A4JUF7 A0A0F7QJW1 A0A3S2PBH0 A0A2W1BHI1 A0A194PDB3 A0A1X9ZM11 A0A0F7QEC7 A0A0L7L438 A0A0N1IAK3 A0A2W1BH31 A0A2H1VEC0 A0A2H1V6B6 A0A194PE80 A0A2A4ITB4 A0A2W1BJJ3 A0A0L7L3S0 A0A2H1VA65 A0A194PCQ5 A0A0N0PDI7 A0A3S2NDD1 A0A2A4ISI6 A0A212FI14 A0A2W1BIE3 A0A2H1V2I4 A0A3S2NTK8 A0A0L7L4E9 A0A2A4IRV9 H9JFH1 A0A1X9ZM28 A0A212EZ25 A0A1B6E8K7 A0A0F7QEA3 A0A194QCB6 A0A2H1V098 A0A2H1VUX4 A0A0N0PF58 A0A0F7QHK5 H9JFX1 A0A2A4ITG3 A0A2J7PXR7 A0A0L7L4A2 A0A2H1VH28 A0A2H1V0B2 A0A2H1VCW8 A0A076FX80 A0A2H1VSX5 A0A2A4JXM9 A0A2A4JB30 H9JFZ3 A0A1B6C5G4 H9JFZ2 A0A1B6I7V1 A0A1B6CXI3 A0A2A4JYR8 A8TUC0 A0A2H1V7P8 A0A076FRA1 H9JFX4 A0A067R2R7 A0A2A4JX39 A0A194QBY5 A0A194PIN2 A0A2C9GUL3 A0A2M4CMZ5 A0A0F7QEA1 A0A1Y9H2G2 A0A1Y9J0A2 W5JEG5 A0A182QVB6 A0A182R8R0 A0A212F791 A0A194QUG5 A0A2A4JY06 A0A182U9S4 A0A1L8DXJ0 A0A0V0G7W2 A0A1B6FB46 A0A1B6JH49 A0A240PL19 A0A182LLK4 A0A194PDZ5 A0A1L8DXS0 A0A3S2NMY0 A0A151X3M2 A0A182JVC0 A0A1L8DXU8 A0A0A9Y5G8 A0A195E3I3 A0A1B6JA43 A0A2M4BAX5 A0A2M4BAZ2 A0A182YKY2 A0A1Y9HAI9
PDB
6Q6Q
E-value=4.8363e-50,
Score=498
Ontologies
GO
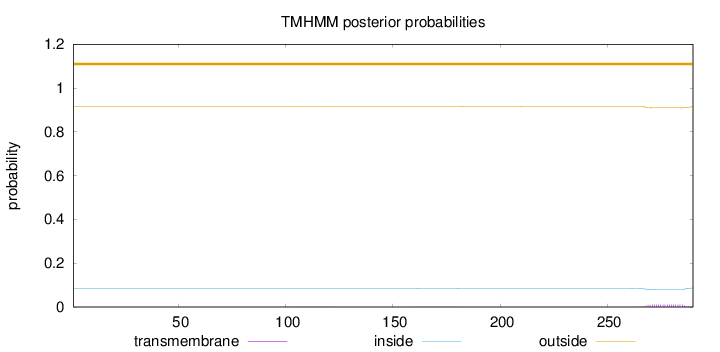
Topology
Length:
290
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.231
Exp number, first 60 AAs:
0.00057
Total prob of N-in:
0.08474
outside
1 - 290
Population Genetic Test Statistics
Pi
267.382898
Theta
208.71386
Tajima's D
0.878987
CLR
0.021667
CSRT
0.632818359082046
Interpretation
Uncertain
