Pre Gene Modal
BGIBMGA008251
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_uncharacterized_protein_LOC101735915_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.056
Sequence
CDS
ATGGGCGACTCACTTTTGAAAGAAGATCCGCTGTCAAGGATCCAGAGAGAGATCATCGAAGTCACAGAGAGGGAAAGAGAATTTAAAGAAGGTCACTTCAGAGTCAACAGGCTTATGTCCGAATCTACGATCGACGTTAATCATCAGAATGGACACATGAACGGCGATGCTGATGAGAAAATATCACGGACCCTCAACAGAGCGGTCTCCATTTCACATTTGTTAGGGCCCATCGCCCCCTCGACTCCTGCAGCGCCTGCTGCCCTCACCGCAAATGGTTACACGCGTCGATTCACTCCGCAGACCGGTACAAAAGGAATAATGCAAAGATTTATAGCCAGCAAAGGAAAGCTGCCCCCGCCTTCAAATGCGGCCTCAACACCGAATACGGCCATCGCCCCTCCCCTTATTTTCCCCTCGACTCCAGTATTGGCACCAGTAGTTTCCCCTCCTATCATCAATCGTACGCCTGAAGGGAAACCAGTACGAAAGGGATACGTCCCCGTCGAAGAGAAGATACAAAAGGAACTACAGGAAATGAAAGACAGAGAACAAGAGCTGAAGAAAATGAGGAAAAAACAGAAACCTTTTGATTTGGAACTGACCGATGATGAGATAAACTCCGAATCAGAAGATGAGGAAATTCCTTTACCCGGGAAACTTAAAGCCACTAAGTCAATAGGAGAGCTTTATGAGGCTCTGAACGAAGACATACGATCGCCATCACCTAGAAATAGTTCCGGACACGACTCACCCGGCAGCCTCAAGCCAGCCAAGTCGCTCGCCGAGCTGTGCGAGCTTGCACCCGGTGAAGAGTTCCTCGCTCCGAGTTCCACTCGGCTCATAAAGCAGTGGGAATCTATGATACAACAGAAACAGGAGGCCGGCGCTGCCTAA
Protein
MGDSLLKEDPLSRIQREIIEVTEREREFKEGHFRVNRLMSESTIDVNHQNGHMNGDADEKISRTLNRAVSISHLLGPIAPSTPAAPAALTANGYTRRFTPQTGTKGIMQRFIASKGKLPPPSNAASTPNTAIAPPLIFPSTPVLAPVVSPPIINRTPEGKPVRKGYVPVEEKIQKELQEMKDREQELKKMRKKQKPFDLELTDDEINSESEDEEIPLPGKLKATKSIGELYEALNEDIRSPSPRNSSGHDSPGSLKPAKSLAELCELAPGEEFLAPSSTRLIKQWESMIQQKQEAGAA
Summary
Uniprot
A0A2H1VS63
A0A1E1W4S1
A0A2W1B982
A0A2A4J731
A0A212FJH7
A0A0L7LK81
+ More
H9JFF4 A0A194PIU4 A0A1E1VXR8 A0A084WNL3 A0A182J1N6 A0A182PD99 A0A182WE43 A0A182MCR2 A0A182FV57 A0A182RXS9 A0A182Y9M1 A0A182T340 W5JD20 A0NBZ8 B0W0L9 A0A1B0ATH4 A0A023EP77 A0A182TWF2 Q17PV9 A0A182UZA4 A0A182HVW2 A0A182WUK9 A0A1I8PUY7 A0A1I8PV10 A0A182KKF0 A0A182N2P0 A0A1L8E8Q5 A0A1L8EG03 A0A1A9ZEM4 A0A1B0FJ81 A0A182QGM7 D3TMC6 A0A1Q3FFF2 A0A1Q3FFD5 A0A1A9VCP0 A0A0L0C6X1 T1PFI5 A0A1I8N4Q6 A0A3B0K1I7 A0A0K8TRW4 A0A3B0JYX0 A0A3B0JTS4 A0A1A9VZ78 A0A1B0DRD5 A0A1A9XBQ8 A0A0K8UA76 A0A0K8UZR7 B3MZ01 B5DKX9 A0A1W4V1M9 B4JNK0 A0A034UZ59 W8B693 A0A1W4WHN9 B4N2G3 B4Q1N1 A0A1Y1MZS2 E2A5R2
H9JFF4 A0A194PIU4 A0A1E1VXR8 A0A084WNL3 A0A182J1N6 A0A182PD99 A0A182WE43 A0A182MCR2 A0A182FV57 A0A182RXS9 A0A182Y9M1 A0A182T340 W5JD20 A0NBZ8 B0W0L9 A0A1B0ATH4 A0A023EP77 A0A182TWF2 Q17PV9 A0A182UZA4 A0A182HVW2 A0A182WUK9 A0A1I8PUY7 A0A1I8PV10 A0A182KKF0 A0A182N2P0 A0A1L8E8Q5 A0A1L8EG03 A0A1A9ZEM4 A0A1B0FJ81 A0A182QGM7 D3TMC6 A0A1Q3FFF2 A0A1Q3FFD5 A0A1A9VCP0 A0A0L0C6X1 T1PFI5 A0A1I8N4Q6 A0A3B0K1I7 A0A0K8TRW4 A0A3B0JYX0 A0A3B0JTS4 A0A1A9VZ78 A0A1B0DRD5 A0A1A9XBQ8 A0A0K8UA76 A0A0K8UZR7 B3MZ01 B5DKX9 A0A1W4V1M9 B4JNK0 A0A034UZ59 W8B693 A0A1W4WHN9 B4N2G3 B4Q1N1 A0A1Y1MZS2 E2A5R2
Pubmed
EMBL
ODYU01003824
SOQ43064.1
GDQN01009077
GDQN01004261
GDQN01000789
JAT81977.1
+ More
JAT86793.1 JAT90265.1 KZ150415 PZC70965.1 NWSH01002900 PCG67334.1 AGBW02008274 OWR53860.1 JTDY01000800 KOB75835.1 BABH01034946 KQ459606 KPI91010.1 GDQN01011528 JAT79526.1 ATLV01024620 KE525353 KFB51807.1 AXCM01001809 AXCM01001810 ADMH02001590 ETN61946.1 AAAB01008823 EAU77464.2 DS231818 EDS41321.1 JXJN01003313 GAPW01002326 JAC11272.1 CH477188 EAT48835.1 APCN01001432 GFDG01003837 JAV14962.1 GFDG01001223 JAV17576.1 CCAG010002766 AXCN02001080 EZ422578 ADD18854.1 GFDL01008768 JAV26277.1 GFDL01008777 JAV26268.1 JRES01000833 KNC27987.1 KA646930 AFP61559.1 OUUW01000003 SPP78881.1 GDAI01000943 JAI16660.1 SPP78879.1 SPP78880.1 AJVK01019925 AJVK01019926 GDHF01028737 GDHF01023968 JAI23577.1 JAI28346.1 GDHF01022984 GDHF01020165 JAI29330.1 JAI32149.1 CH902632 EDV32845.1 CH379063 EDY72179.2 CH916371 EDV92293.1 GAKP01023394 JAC35564.1 GAMC01009870 JAB96685.1 CH963925 EDW78552.1 CM000162 EDX01472.1 GEZM01017461 GEZM01017460 JAV90658.1 GL437023 EFN71259.1
JAT86793.1 JAT90265.1 KZ150415 PZC70965.1 NWSH01002900 PCG67334.1 AGBW02008274 OWR53860.1 JTDY01000800 KOB75835.1 BABH01034946 KQ459606 KPI91010.1 GDQN01011528 JAT79526.1 ATLV01024620 KE525353 KFB51807.1 AXCM01001809 AXCM01001810 ADMH02001590 ETN61946.1 AAAB01008823 EAU77464.2 DS231818 EDS41321.1 JXJN01003313 GAPW01002326 JAC11272.1 CH477188 EAT48835.1 APCN01001432 GFDG01003837 JAV14962.1 GFDG01001223 JAV17576.1 CCAG010002766 AXCN02001080 EZ422578 ADD18854.1 GFDL01008768 JAV26277.1 GFDL01008777 JAV26268.1 JRES01000833 KNC27987.1 KA646930 AFP61559.1 OUUW01000003 SPP78881.1 GDAI01000943 JAI16660.1 SPP78879.1 SPP78880.1 AJVK01019925 AJVK01019926 GDHF01028737 GDHF01023968 JAI23577.1 JAI28346.1 GDHF01022984 GDHF01020165 JAI29330.1 JAI32149.1 CH902632 EDV32845.1 CH379063 EDY72179.2 CH916371 EDV92293.1 GAKP01023394 JAC35564.1 GAMC01009870 JAB96685.1 CH963925 EDW78552.1 CM000162 EDX01472.1 GEZM01017461 GEZM01017460 JAV90658.1 GL437023 EFN71259.1
Proteomes
UP000218220
UP000007151
UP000037510
UP000005204
UP000053268
UP000030765
+ More
UP000075880 UP000075885 UP000075920 UP000075883 UP000069272 UP000075900 UP000076408 UP000075901 UP000000673 UP000007062 UP000002320 UP000092460 UP000075902 UP000008820 UP000075903 UP000075840 UP000076407 UP000095300 UP000075882 UP000075884 UP000092445 UP000092444 UP000075886 UP000078200 UP000037069 UP000095301 UP000268350 UP000091820 UP000092462 UP000092443 UP000007801 UP000001819 UP000192221 UP000001070 UP000192223 UP000007798 UP000002282 UP000000311
UP000075880 UP000075885 UP000075920 UP000075883 UP000069272 UP000075900 UP000076408 UP000075901 UP000000673 UP000007062 UP000002320 UP000092460 UP000075902 UP000008820 UP000075903 UP000075840 UP000076407 UP000095300 UP000075882 UP000075884 UP000092445 UP000092444 UP000075886 UP000078200 UP000037069 UP000095301 UP000268350 UP000091820 UP000092462 UP000092443 UP000007801 UP000001819 UP000192221 UP000001070 UP000192223 UP000007798 UP000002282 UP000000311
PRIDE
Pfam
PF15304 AKAP2_C
Interpro
IPR029304
AKAP2_C
ProteinModelPortal
A0A2H1VS63
A0A1E1W4S1
A0A2W1B982
A0A2A4J731
A0A212FJH7
A0A0L7LK81
+ More
H9JFF4 A0A194PIU4 A0A1E1VXR8 A0A084WNL3 A0A182J1N6 A0A182PD99 A0A182WE43 A0A182MCR2 A0A182FV57 A0A182RXS9 A0A182Y9M1 A0A182T340 W5JD20 A0NBZ8 B0W0L9 A0A1B0ATH4 A0A023EP77 A0A182TWF2 Q17PV9 A0A182UZA4 A0A182HVW2 A0A182WUK9 A0A1I8PUY7 A0A1I8PV10 A0A182KKF0 A0A182N2P0 A0A1L8E8Q5 A0A1L8EG03 A0A1A9ZEM4 A0A1B0FJ81 A0A182QGM7 D3TMC6 A0A1Q3FFF2 A0A1Q3FFD5 A0A1A9VCP0 A0A0L0C6X1 T1PFI5 A0A1I8N4Q6 A0A3B0K1I7 A0A0K8TRW4 A0A3B0JYX0 A0A3B0JTS4 A0A1A9VZ78 A0A1B0DRD5 A0A1A9XBQ8 A0A0K8UA76 A0A0K8UZR7 B3MZ01 B5DKX9 A0A1W4V1M9 B4JNK0 A0A034UZ59 W8B693 A0A1W4WHN9 B4N2G3 B4Q1N1 A0A1Y1MZS2 E2A5R2
H9JFF4 A0A194PIU4 A0A1E1VXR8 A0A084WNL3 A0A182J1N6 A0A182PD99 A0A182WE43 A0A182MCR2 A0A182FV57 A0A182RXS9 A0A182Y9M1 A0A182T340 W5JD20 A0NBZ8 B0W0L9 A0A1B0ATH4 A0A023EP77 A0A182TWF2 Q17PV9 A0A182UZA4 A0A182HVW2 A0A182WUK9 A0A1I8PUY7 A0A1I8PV10 A0A182KKF0 A0A182N2P0 A0A1L8E8Q5 A0A1L8EG03 A0A1A9ZEM4 A0A1B0FJ81 A0A182QGM7 D3TMC6 A0A1Q3FFF2 A0A1Q3FFD5 A0A1A9VCP0 A0A0L0C6X1 T1PFI5 A0A1I8N4Q6 A0A3B0K1I7 A0A0K8TRW4 A0A3B0JYX0 A0A3B0JTS4 A0A1A9VZ78 A0A1B0DRD5 A0A1A9XBQ8 A0A0K8UA76 A0A0K8UZR7 B3MZ01 B5DKX9 A0A1W4V1M9 B4JNK0 A0A034UZ59 W8B693 A0A1W4WHN9 B4N2G3 B4Q1N1 A0A1Y1MZS2 E2A5R2
Ontologies
GO
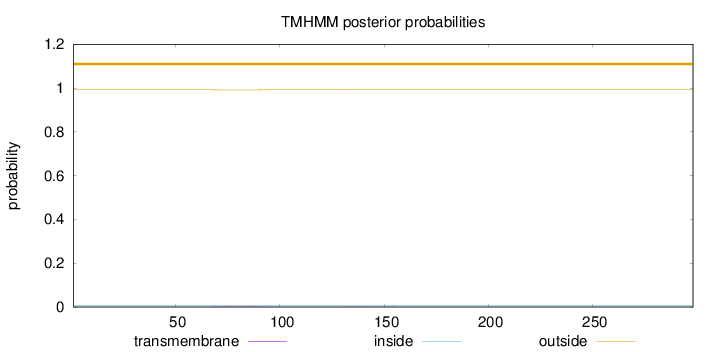
Topology
Length:
298
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06929
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00822
outside
1 - 298
Population Genetic Test Statistics
Pi
254.005285
Theta
178.833269
Tajima's D
0.954206
CLR
0.26602
CSRT
0.646667666616669
Interpretation
Uncertain
