Gene
KWMTBOMO10660
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.105
Sequence
CDS
ATGGACTTCGAGTCCGCGACTAGCCCTCAACCCGGAACCTCGGATGGGTTCCAAACTGTAACGCGCGGTAAAAAGCGTACTCGCGTCGTGGAGTCCCGGAGCTCCACGACGAAGCAAACCAAGTCCGCGACCGCCTCCCGACCGCAGGTAGTCGTAACGCCGGAGTCGGACTCCGCCCGCCGCGTAATCCCACCGCCGCGCCCAAAAACCGCGCCCGCGCCTAAAACAGTTGTCCCGCCCCCGCTGATACTCCAGGAGAAGTCAGCGTGGAATCGCGTATCCCAGGCCCTTCAGGCCAACAAAATTAATTATACCCATGCGCGTAACGTCGCGCATGGGATTCAGATTAAGGTCGCAACGCCGGGCGACCATAGGGCCCTCTCTGCTTACCTCCGAAAGGAGAACATAGGTTATCACACCTATGCTCTTCAGGAGGACCGCGAACTCCGCGTAGTGATACGTGGAGTCCCCAAGGAACTCGACATAGACTACGTAAAGGAGGATCTGATCGGTCAGTCCCTCCCAATAATTAGTGTGCACCGGATGCACAGCGGACGCGGCAGACAGCCGTACAATATGATTCTCGTCGCGCTAGAATCAACCCCGGAGGCAAAGAAAAGAATCTCATGTCTCAAAACGATATGCGGCCTCTCCGGGGTCACCATCGAAGCTCCCCATAAACGTGGTACTCCCGGGCAGTGCCACAGGTGCCAGCTCTATGGCCACTCAGCGCGTAATTGCCACGCGCGCCCCCGCTGCGTGAAATGCCTCGGCGATCACGCAACCACAGAATGTTCGCGTGTTAGGGAAACCGCGACGGAACCCCCAAGCTGTGTCCTATGCCTTAAGCAAGGGCACCCGGCAAACTACCGCGGATGTCCTAGGGCTCCGCGTAAACGCCTGAACCACCCAGCCCCCCGAACCGTCGACCCAAAGACTTCGGCGCCTTCAGTGCCGAAACCAGCCTTCGTACCGGCTGCGGTTCCCACCGTCTCGGCGTGGAAAAAACCGCTGCCGTATACGAAGGAGGGAACACAAAACGTACCCCAGCTCCCGCCTGCGACACGTCACGCGCCTCAGCCCCTGCTCGCACCTCGCCCCGCGCCCGCGTACCGCCCCCCGCAACCCTCAGTCGTCAACGACTTCGCGCTCGTGCGCGACTTCGTCACCGCGGTAAACTTTGACCGTCTGCGATCGTTCGCAGATGCTATCCGTAGATCTTCAAAAGAAGTCGATGTGGAGGACAGCTTCCACCGCATCAGACTGTCCTCCGAACTTAGGAATCTCTTAAGAGTTAGGAACGCGGCAATCCGGGCCTACGATCGTCTTCCCACGCATTCAAACCGGATTCGGATGCGTCGTCTACAACGCGAAGTCCACTCCCGCCTAAGTGACGCGCGTAACGATAATTGGCATAGTTATTTAGAACAACTCGCGCCCTCCCACCAAGCATACTGGCGACTAGCTAGGACTCTCAAATCCGAAACTACCGCTACTATGCCTCCCCTCGTACGCCCTTCAGGCCAACCACCGGCATTCGATGACGATGACAAGGCTGAGCTGCTGGCCGATGCACTGCAAGAGCAGTGCACCACCAGCACTCAACACGCGGACCCCGAACACACCGAGTTAGTCGACAGGGAGGTCGAGCGCAGAGCTTCCCTGCCGCCCTCGGACGCGTTACCCCCCATTACCACTGACGAAGTTAGAGACGCGATCCACAACCTCCAACCTAGGAAGGCACCCGGCTCCGACGGCATCCGCAACCGCGCGCTAAAACTCTTGCCAGTCCAACTGATAGCAATGTTGGCTACAATTTTAAATGCCGCTATGACGCACTGCATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGTATACATAAGCCGGGCAAACCGACAAACGAAACTTCTAGTTACCGTCCGATTAGTCTCCTCCCGACGATAGGAAAAATTTACGAACGGCTTCTTAGGAAACGCCTCTGGGATTTTGTTACCGCGAACAAAATTCTCATAGACGAACAGTTCGGATTCCGCTCTAAACACTCGTGCGTACAACAAGTGCACCGCCTCACGGAGCACATCCTGATAGGACTAAATAGGCGTAAACAAATCCCGACCGGCGCCCTCTTCTTCGACATCGCGAAGGCGTTCGACAAAGTCTGGCACAACGGTTTAATTTACAAACTGTACAACATGGGAGTGCCAGACAGACTCGTGCTCATCATACGAGACTTCTTGTCGAACCGTTCGTTTCGATATCGAGTAGAGGGAACTCGTTCTCGTCCCCGTCAACTGACTGCCGGAGTCCCGCAAGGCTCCGCGCTCTCCCCGTTATTATTTAGTTTGTATATCAATGATATACCCCGGTCTCCGGAGACCCATCTAGCGCTCTTCGCCGATGACACGGCTATCTACTACTCGTGTAGGAAGATGTCGCTGCTTCATCGGCGACTCCAGATCGCAGTAGCCACCATGGGACAGTGGTTCCGGAAGTGGCGCATCGACATCAACCCCACGAAAAGCACAGCGGTGCTCTTCAAAAGGGGTCGCCCTCCGAATACCACTTCGAGCATCCCACTCCCTAATAGGCGCGCAAACACCTCCGCCGTTAGCCCCGTCACTCTCTTTGGCCAGCCCATACCGTGGGCCTCGCAGGTCAAATACCTAGGCGTCACCCTCGACAGAGGGATGACATTCCGTCCCCATATTAAAACGGTACGCGACCGTGCCGCCTTCATATTAGGACGTCTCTACCCTATGCTTTGCAAGCGAAGCAAACTGTCCCTCCGTAATAAGGTAACTCTCTACAAAACTTGCATACGCCCCGTTATGACGTATGCAAGCGTAGTGTTCGCTCACGCAGCCCGCACCAACTTGAAGCCCCTTCAGGTTATTCAATCCCGATTCTGCAGGATAGCCGTCGGAGCACCATGGTTCCTGAGGAACGTGGATGTCCACGATGACCTGGAGCTTGACTCTGTCAGTAAGTATCTACAGTCGGCATCGCTGCGCCATTTTGAGAAGGCGGCACGACATGAGAACCCTCTCATCGTAGCCGCTGGAAATTACATACCCGACCCAGTAGACCGAATGGTAAACCGTCGACGTCGCCCAAAGCACGTCATTACGGATCCTCCTGATCCATTAACGGTGCTCTTAGGCACCACAAGCACCGGTCACCGTCCTCGTCGAACCCGTCGCTTGCGACGAAGGGCTCGACGAGCGAACTAA
Protein
MDFESATSPQPGTSDGFQTVTRGKKRTRVVESRSSTTKQTKSATASRPQVVVTPESDSARRVIPPPRPKTAPAPKTVVPPPLILQEKSAWNRVSQALQANKINYTHARNVAHGIQIKVATPGDHRALSAYLRKENIGYHTYALQEDRELRVVIRGVPKELDIDYVKEDLIGQSLPIISVHRMHSGRGRQPYNMILVALESTPEAKKRISCLKTICGLSGVTIEAPHKRGTPGQCHRCQLYGHSARNCHARPRCVKCLGDHATTECSRVRETATEPPSCVLCLKQGHPANYRGCPRAPRKRLNHPAPRTVDPKTSAPSVPKPAFVPAAVPTVSAWKKPLPYTKEGTQNVPQLPPATRHAPQPLLAPRPAPAYRPPQPSVVNDFALVRDFVTAVNFDRLRSFADAIRRSSKEVDVEDSFHRIRLSSELRNLLRVRNAAIRAYDRLPTHSNRIRMRRLQREVHSRLSDARNDNWHSYLEQLAPSHQAYWRLARTLKSETTATMPPLVRPSGQPPAFDDDDKAELLADALQEQCTTSTQHADPEHTELVDREVERRASLPPSDALPPITTDEVRDAIHNLQPRKAPGSDGIRNRALKLLPVQLIAMLATILNAAMTHCIFPAVWKEADVIGIHKPGKPTNETSSYRPISLLPTIGKIYERLLRKRLWDFVTANKILIDEQFGFRSKHSCVQQVHRLTEHILIGLNRRKQIPTGALFFDIAKAFDKVWHNGLIYKLYNMGVPDRLVLIIRDFLSNRSFRYRVEGTRSRPRQLTAGVPQGSALSPLLFSLYINDIPRSPETHLALFADDTAIYYSCRKMSLLHRRLQIAVATMGQWFRKWRIDINPTKSTAVLFKRGRPPNTTSSIPLPNRRANTSAVSPVTLFGQPIPWASQVKYLGVTLDRGMTFRPHIKTVRDRAAFILGRLYPMLCKRSKLSLRNKVTLYKTCIRPVMTYASVVFAHAARTNLKPLQVIQSRFCRIAVGAPWFLRNVDVHDDLELDSVSKYLQSASLRHFEKAARHENPLIVAAGNYIPDPVDRMVNRRRRPKHVITDPPDPLTVLLGTTSTGHRPRRTRRLRRRARRAN
Summary
Uniprot
ProteinModelPortal
PDB
6AR3
E-value=0.00312712,
Score=100
Ontologies
GO
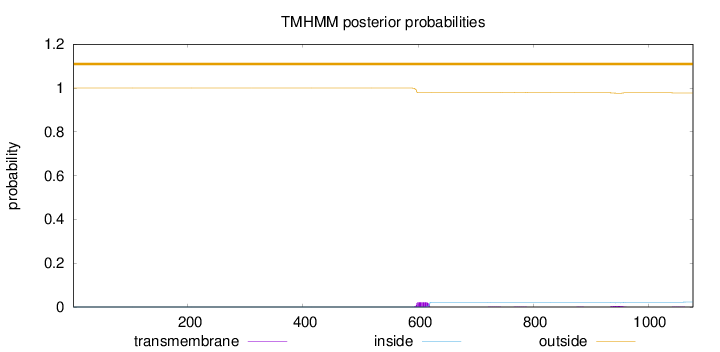
Topology
Length:
1078
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.623199999999998
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00037
outside
1 - 1078
Population Genetic Test Statistics
Pi
0.174054
Theta
0.174643
Tajima's D
0
CLR
20.347284
CSRT
0.245187740612969
Interpretation
Uncertain
