Gene
KWMTBOMO10658
Pre Gene Modal
BGIBMGA008257
Annotation
PREDICTED:_SLIT-ROBO_Rho_GTPase-activating_protein_3-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.077 Mitochondrial Reliability : 1.545 Nuclear Reliability : 1.492
Sequence
CDS
ATGCGGGAGGGGCGCGGAGGGCGAATCCCGCGCGGAGGCTGCGCGACCTCAGTCGAGCGCCGCCGCTCCGCCATGTTCCACTGCATCGTGCAGCCGCGGCATGACTGGGGACCGCCAGACTTCGCCAATGCCTTCCCCGATATAAAGTCACAACTAGTGGAGCACACTCGCGTGCTAGAGGCTCGCGCGGAGGCGACGGCGGGCGTGGCTGGGGAACTCCACGAGTACTGCCGGAGACGCGCCGACCTGGAACACGAGTACGCGAGGGCACTCGACAAGCTCGCCCGCGCCGCGCACCAGCGACACGAGGGACAGAAACACAAACGCGAGCAATGGGCGATCACGGGCGCGTTCTCGTGCTGGCAGACGGCGCTGGACGCGACGCGCTCGCTGTCCCGGGACCACGCCGCGCTCGCCGACCTGTACGGGGGCCCGCTGGCCGCGCGCCTGCAGCGGGCCGCCGACGACGCGCTCCGCCTGCACAGGAAGTGCCGGGACATCGTGTCTGAGCGGCACGAGGAGGTGTGCGCGGCGCTGGCGGAGGCGTGGGGCGCGGGCAAGGCGCAGACCGCCGCGGCGCACGAGTGGCGCGTCGCCGCACACAAGCTGCGCACCGCACACGCCGCGCGCGCCGCCCTCGCCGCACACTCGCCGCCGCGACACAAGAAGCTCAAGGCGCTCGACAAGGAACTAGACAAGCGTCGCTCGCGGCACAGCGCGGCGCGCGCACACGCGCTGCGGGCCCGCGCCGACTACGTGCTCAGCCTCGAGGCCGCCAACGCCACGCTGCAGCGGTACTTCCTCGACGACATCGCCGACATCATCCTGTGCATGGAGGTGGGGTTCGAGGCGGTGGTGGGGCGCGCCGTGCGCACGCTGGGCGCGGCGGAGGGCGGCGCGGGGCGGGCGCGGGCGGGCGCGGGGGAGGCGCTGTGCGGCGCGGCGGCGGCGCTGGACGCGCTGGCGGACCGGCGCCGCCTGCTGGACGCGCACGTGGCCGCCTTCGCGCTGCCCCGCCCGCTGCCGCACCAGCCGCCGCCGCCGGACAGTTAA
Protein
MREGRGGRIPRGGCATSVERRRSAMFHCIVQPRHDWGPPDFANAFPDIKSQLVEHTRVLEARAEATAGVAGELHEYCRRRADLEHEYARALDKLARAAHQRHEGQKHKREQWAITGAFSCWQTALDATRSLSRDHAALADLYGGPLAARLQRAADDALRLHRKCRDIVSERHEEVCAALAEAWGAGKAQTAAAHEWRVAAHKLRTAHAARAALAAHSPPRHKKLKALDKELDKRRSRHSAARAHALRARADYVLSLEAANATLQRYFLDDIADIILCMEVGFEAVVGRAVRTLGAAEGGAGRARAGAGEALCGAAAALDALADRRRLLDAHVAAFALPRPLPHQPPPPDS
Summary
Uniprot
H9JFG0
A0A3S2N953
A0A212F2J5
A0A1B6DVJ2
A0A1B6DN07
Q7Q2Q9
+ More
A0A336KVU5 A0A1J1HGH2 A0A182YI20 A0A182NND1 A0A182WYK0 A0A182RXF5 A0A182P8D3 A0A182QKV9 A0A182WCM4 A0A1J1HHD6 A0A336KSW8 A0A182FLQ9 A0A2M4B9E2 A0A182K696 A0A139WNS8 A0A1B0FV14 A0A2M4CU92 A0A0P4WQA1 A0A154NWY6 A0A0C9RYJ4 A0A0C9QI22 K7IRA6 A0A1B6E4U3 E2BUE0 A0A0L7QZU7 A0A1B6DHB9 A0A088A2B7 A0A067RG87 A0A1A9VZE0 A0A232ETF0 A0A2A3EBX1 V9IC58 A0A1B0ESN9 A0A1A9UQ51 A0A1B0A8V4 A0A1A9XMX8 A0A1B0BZ76 A0A0J7L0N9 A0A026W0C0 A0A3L8D5V0 A0A182SWI1 J9KA86 A0A195AT22 E0VFJ7 A0A1J1HL54 A0A182HXJ3 A0A1B6G5P1 A0A1I8MLH9 A0A1Q3FW79 A0A1Q3FW69 D6W740 A0A1I8PXD9 A0A1B6EIP6 B0X4W0 A0A1Y1LKP1 A0A2M4CUZ9 A0A2M4CUW8 A0A2M4A9L8 A0A182MER2 A0A2M4B9J9 A0A2M4B9K6 A0A2M3Z406 A0A2P8XLE4 A0A182GVW6 A0A0P4WHY1 A0A0P4WXW8 A0A0P4WHM4 A0A0M9A465
A0A336KVU5 A0A1J1HGH2 A0A182YI20 A0A182NND1 A0A182WYK0 A0A182RXF5 A0A182P8D3 A0A182QKV9 A0A182WCM4 A0A1J1HHD6 A0A336KSW8 A0A182FLQ9 A0A2M4B9E2 A0A182K696 A0A139WNS8 A0A1B0FV14 A0A2M4CU92 A0A0P4WQA1 A0A154NWY6 A0A0C9RYJ4 A0A0C9QI22 K7IRA6 A0A1B6E4U3 E2BUE0 A0A0L7QZU7 A0A1B6DHB9 A0A088A2B7 A0A067RG87 A0A1A9VZE0 A0A232ETF0 A0A2A3EBX1 V9IC58 A0A1B0ESN9 A0A1A9UQ51 A0A1B0A8V4 A0A1A9XMX8 A0A1B0BZ76 A0A0J7L0N9 A0A026W0C0 A0A3L8D5V0 A0A182SWI1 J9KA86 A0A195AT22 E0VFJ7 A0A1J1HL54 A0A182HXJ3 A0A1B6G5P1 A0A1I8MLH9 A0A1Q3FW79 A0A1Q3FW69 D6W740 A0A1I8PXD9 A0A1B6EIP6 B0X4W0 A0A1Y1LKP1 A0A2M4CUZ9 A0A2M4CUW8 A0A2M4A9L8 A0A182MER2 A0A2M4B9J9 A0A2M4B9K6 A0A2M3Z406 A0A2P8XLE4 A0A182GVW6 A0A0P4WHY1 A0A0P4WXW8 A0A0P4WHM4 A0A0M9A465
Pubmed
EMBL
BABH01034930
RSAL01000519
RVE41466.1
AGBW02010729
OWR47965.1
GEDC01007593
+ More
JAS29705.1 GEDC01010249 JAS27049.1 AAAB01008968 EAA13247.4 UFQS01001054 UFQT01001054 SSX08742.1 SSX28654.1 CVRI01000002 CRK86971.1 AXCN02001907 CRK86970.1 UFQS01000969 UFQT01000969 SSX08087.1 SSX28232.1 GGFJ01000533 MBW49674.1 KQ971307 KYB29649.1 AJWK01006530 AJWK01006531 AJWK01006532 AJWK01006533 AJWK01006534 AJWK01006535 GGFL01004736 MBW68914.1 GDRN01050209 JAI66483.1 KQ434775 KZC04072.1 GBYB01014340 JAG84107.1 GBYB01014343 JAG84110.1 GEDC01004357 JAS32941.1 GL450640 EFN80667.1 KQ414673 KOC64135.1 GEDC01012225 JAS25073.1 KK852486 KDR22787.1 NNAY01002280 OXU21638.1 KZ288311 PBC28541.1 JR037675 JR037676 AEY57909.1 AEY57910.1 CCAG010023381 JXJN01023024 LBMM01001415 KMQ96362.1 KK107599 EZA48584.1 QOIP01000012 RLU15857.1 ABLF02039578 KQ976745 KYM75368.1 AAZO01001856 DS235114 EEB12153.1 CRK86969.1 APCN01005314 GECZ01012136 JAS57633.1 GFDL01003196 JAV31849.1 GFDL01003191 JAV31854.1 EFA11413.2 GECZ01031983 JAS37786.1 DS232356 EDS40575.1 GEZM01059008 JAV71577.1 GGFL01004965 MBW69143.1 GGFL01004966 MBW69144.1 GGFK01004172 MBW37493.1 AXCM01003886 GGFJ01000566 MBW49707.1 GGFJ01000565 MBW49706.1 GGFM01002484 MBW23235.1 PYGN01001794 PSN32784.1 JXUM01091911 JXUM01091912 JXUM01091913 JXUM01091914 JXUM01091915 JXUM01091916 KQ563951 KXJ73026.1 GDRN01050211 JAI66481.1 GDRN01050210 JAI66482.1 GDRN01050208 JAI66484.1 KQ435762 KOX75499.1
JAS29705.1 GEDC01010249 JAS27049.1 AAAB01008968 EAA13247.4 UFQS01001054 UFQT01001054 SSX08742.1 SSX28654.1 CVRI01000002 CRK86971.1 AXCN02001907 CRK86970.1 UFQS01000969 UFQT01000969 SSX08087.1 SSX28232.1 GGFJ01000533 MBW49674.1 KQ971307 KYB29649.1 AJWK01006530 AJWK01006531 AJWK01006532 AJWK01006533 AJWK01006534 AJWK01006535 GGFL01004736 MBW68914.1 GDRN01050209 JAI66483.1 KQ434775 KZC04072.1 GBYB01014340 JAG84107.1 GBYB01014343 JAG84110.1 GEDC01004357 JAS32941.1 GL450640 EFN80667.1 KQ414673 KOC64135.1 GEDC01012225 JAS25073.1 KK852486 KDR22787.1 NNAY01002280 OXU21638.1 KZ288311 PBC28541.1 JR037675 JR037676 AEY57909.1 AEY57910.1 CCAG010023381 JXJN01023024 LBMM01001415 KMQ96362.1 KK107599 EZA48584.1 QOIP01000012 RLU15857.1 ABLF02039578 KQ976745 KYM75368.1 AAZO01001856 DS235114 EEB12153.1 CRK86969.1 APCN01005314 GECZ01012136 JAS57633.1 GFDL01003196 JAV31849.1 GFDL01003191 JAV31854.1 EFA11413.2 GECZ01031983 JAS37786.1 DS232356 EDS40575.1 GEZM01059008 JAV71577.1 GGFL01004965 MBW69143.1 GGFL01004966 MBW69144.1 GGFK01004172 MBW37493.1 AXCM01003886 GGFJ01000566 MBW49707.1 GGFJ01000565 MBW49706.1 GGFM01002484 MBW23235.1 PYGN01001794 PSN32784.1 JXUM01091911 JXUM01091912 JXUM01091913 JXUM01091914 JXUM01091915 JXUM01091916 KQ563951 KXJ73026.1 GDRN01050211 JAI66481.1 GDRN01050210 JAI66482.1 GDRN01050208 JAI66484.1 KQ435762 KOX75499.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000007062
UP000183832
UP000076408
+ More
UP000075884 UP000076407 UP000075900 UP000075885 UP000075886 UP000075920 UP000069272 UP000075881 UP000007266 UP000092461 UP000076502 UP000002358 UP000008237 UP000053825 UP000005203 UP000027135 UP000091820 UP000215335 UP000242457 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000036403 UP000053097 UP000279307 UP000075901 UP000007819 UP000078540 UP000009046 UP000075840 UP000095301 UP000095300 UP000002320 UP000075883 UP000245037 UP000069940 UP000249989 UP000053105
UP000075884 UP000076407 UP000075900 UP000075885 UP000075886 UP000075920 UP000069272 UP000075881 UP000007266 UP000092461 UP000076502 UP000002358 UP000008237 UP000053825 UP000005203 UP000027135 UP000091820 UP000215335 UP000242457 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000036403 UP000053097 UP000279307 UP000075901 UP000007819 UP000078540 UP000009046 UP000075840 UP000095301 UP000095300 UP000002320 UP000075883 UP000245037 UP000069940 UP000249989 UP000053105
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JFG0
A0A3S2N953
A0A212F2J5
A0A1B6DVJ2
A0A1B6DN07
Q7Q2Q9
+ More
A0A336KVU5 A0A1J1HGH2 A0A182YI20 A0A182NND1 A0A182WYK0 A0A182RXF5 A0A182P8D3 A0A182QKV9 A0A182WCM4 A0A1J1HHD6 A0A336KSW8 A0A182FLQ9 A0A2M4B9E2 A0A182K696 A0A139WNS8 A0A1B0FV14 A0A2M4CU92 A0A0P4WQA1 A0A154NWY6 A0A0C9RYJ4 A0A0C9QI22 K7IRA6 A0A1B6E4U3 E2BUE0 A0A0L7QZU7 A0A1B6DHB9 A0A088A2B7 A0A067RG87 A0A1A9VZE0 A0A232ETF0 A0A2A3EBX1 V9IC58 A0A1B0ESN9 A0A1A9UQ51 A0A1B0A8V4 A0A1A9XMX8 A0A1B0BZ76 A0A0J7L0N9 A0A026W0C0 A0A3L8D5V0 A0A182SWI1 J9KA86 A0A195AT22 E0VFJ7 A0A1J1HL54 A0A182HXJ3 A0A1B6G5P1 A0A1I8MLH9 A0A1Q3FW79 A0A1Q3FW69 D6W740 A0A1I8PXD9 A0A1B6EIP6 B0X4W0 A0A1Y1LKP1 A0A2M4CUZ9 A0A2M4CUW8 A0A2M4A9L8 A0A182MER2 A0A2M4B9J9 A0A2M4B9K6 A0A2M3Z406 A0A2P8XLE4 A0A182GVW6 A0A0P4WHY1 A0A0P4WXW8 A0A0P4WHM4 A0A0M9A465
A0A336KVU5 A0A1J1HGH2 A0A182YI20 A0A182NND1 A0A182WYK0 A0A182RXF5 A0A182P8D3 A0A182QKV9 A0A182WCM4 A0A1J1HHD6 A0A336KSW8 A0A182FLQ9 A0A2M4B9E2 A0A182K696 A0A139WNS8 A0A1B0FV14 A0A2M4CU92 A0A0P4WQA1 A0A154NWY6 A0A0C9RYJ4 A0A0C9QI22 K7IRA6 A0A1B6E4U3 E2BUE0 A0A0L7QZU7 A0A1B6DHB9 A0A088A2B7 A0A067RG87 A0A1A9VZE0 A0A232ETF0 A0A2A3EBX1 V9IC58 A0A1B0ESN9 A0A1A9UQ51 A0A1B0A8V4 A0A1A9XMX8 A0A1B0BZ76 A0A0J7L0N9 A0A026W0C0 A0A3L8D5V0 A0A182SWI1 J9KA86 A0A195AT22 E0VFJ7 A0A1J1HL54 A0A182HXJ3 A0A1B6G5P1 A0A1I8MLH9 A0A1Q3FW79 A0A1Q3FW69 D6W740 A0A1I8PXD9 A0A1B6EIP6 B0X4W0 A0A1Y1LKP1 A0A2M4CUZ9 A0A2M4CUW8 A0A2M4A9L8 A0A182MER2 A0A2M4B9J9 A0A2M4B9K6 A0A2M3Z406 A0A2P8XLE4 A0A182GVW6 A0A0P4WHY1 A0A0P4WXW8 A0A0P4WHM4 A0A0M9A465
PDB
5I6R
E-value=5.15923e-09,
Score=145
Ontologies
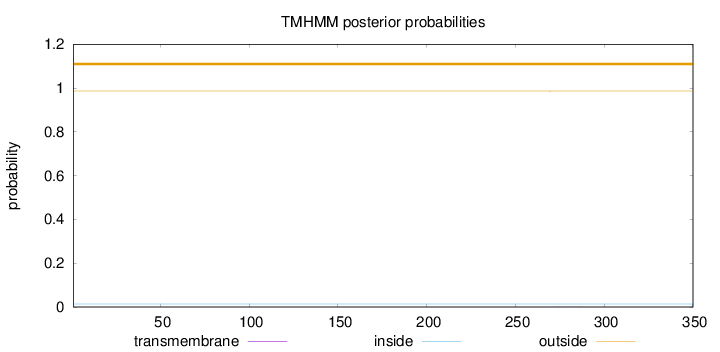
Topology
Length:
350
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00497
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01383
outside
1 - 350
Population Genetic Test Statistics
Pi
171.784852
Theta
146.299924
Tajima's D
0.5838
CLR
0.086606
CSRT
0.543022848857557
Interpretation
Uncertain
