Gene
KWMTBOMO10655
Pre Gene Modal
BGIBMGA008259
Annotation
PREDICTED:_regulator_of_G-protein_signaling_20_isoform_X3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.407 Nuclear Reliability : 1.8
Sequence
CDS
ATGGCAAGTAGTCAGATTAGTCTGGCGGCGCGCGGCTCGGAGGCCGGCCCCGGCAAGAAGCCTCCCGAACCGGGACCACAGGAACCCCTTCTACAGGAAGGCGACGCGCCGCCATCGCTGGAGGAGATCCAAGGCTGGGGGCAGTCGTTCGACCGGCTGATGCGGAGCACCGCCGGCAGGAAGGTGTTTCGTGACTTCCTGCGCGGCGAGTACTCGGAGGAGAACATAATGTTCTGGCTCGCGTGCGAAGAGCTCAAGCGAGAGACCGACCCCGACGCCGTCGAGGAGAAGGCGCGCTTTATCTACGAGGACTACATCTCTATACTGTCCCCTAAGGAGGTGTCTCTCGACTCGCGGGTCCGAGAGATCGTGAACCGCAACATGGTGGAGCCCACGCCGCACACGTTCGACGAGGCGCAGCTGCAGATCTACACGCTGATGCACCGCGACTCGTACCCGCGGTTCGTCAACTCGCCGCTCTACAAGACGCTGGCGCGGCTGCCGGAGCCGTGA
Protein
MASSQISLAARGSEAGPGKKPPEPGPQEPLLQEGDAPPSLEEIQGWGQSFDRLMRSTAGRKVFRDFLRGEYSEENIMFWLACEELKRETDPDAVEEKARFIYEDYISILSPKEVSLDSRVREIVNRNMVEPTPHTFDEAQLQIYTLMHRDSYPRFVNSPLYKTLARLPEP
Summary
Uniprot
A0A3S2TBZ1
A0A194PDE7
A0A212F2M3
A0A2A4JIQ1
A0A2A4JJ59
A0A182RP65
+ More
A0A084WK75 A0A182FC83 A0A182M6Q9 A0A182NK54 A0A182YEL5 A0A182T0R3 A0A182P0N8 A0A182WLI8 A0A182QDH1 A0A1D6T4Q2 A0A182KQE1 A0A182WWZ6 A0A182V695 A0A182I2J3 A0A182TH05 Q7QIY2 B0WC38 A0A1B6LIX0 A0A1B6C0T6 W5JHN4 A0A336MT64 A0A1Y1KR19 A0A1E1X245 A0A1E1XJY5 T1HZN2 A0A336MRP1 A0A139WPB2 A0A0T6B1P3 A0A1D2NCY7 A0A1W4XK38 A0A1J1JA06 A0A147BKU8 L7MCQ3 A0A182G712 A0A0J7LC56 E2BDJ3 A0A151X9T4 A0A151J3M5 A0A0K8WD88 A0A034WX38 A0A195BEX0 A0A0P6CWD0 A0A0P5SE32 A0A2R7WDR8 A0A162CZK0 A0A0J9U4W3 B4HN86 B4QBR7 Q8T017 A0A0B4LFL6 A0A158NBW6 A0A0A1X3L8 F4W5Q0 A0A0K8U7K7 B4P527 A0A195FLX4 A0A1I8PQJ7 W8ANF3 B3NLK8 E2AKB2 A0A0Q9WEJ6 A0A0A1WRB4 A0A0P4WWE9 A0A1W4V0V5 B4LKT0 B4MYI6 S4RT90 A0A210PTU2 B4J9K2 B4KQU7 A0A226EZ15 R7UET2 A0A1S3KI58 B3MHX5 A0A2C9JVV8 A0A1B0G4D5 Q28XE9 A0A1A9ZNI8 B4GIU8 A0A1A9VIH7 A0A195C9X0 A0A0M3QVF5 A0A3B0J1Q1 V4B1L7 A0A1I8NAB1 A0A1A9XJ55 A0A1A9X3X0 E0VFJ5 A0A2A3ECN6 V9IEC3 A0A088A5Y3 A0A3M7R5K5
A0A084WK75 A0A182FC83 A0A182M6Q9 A0A182NK54 A0A182YEL5 A0A182T0R3 A0A182P0N8 A0A182WLI8 A0A182QDH1 A0A1D6T4Q2 A0A182KQE1 A0A182WWZ6 A0A182V695 A0A182I2J3 A0A182TH05 Q7QIY2 B0WC38 A0A1B6LIX0 A0A1B6C0T6 W5JHN4 A0A336MT64 A0A1Y1KR19 A0A1E1X245 A0A1E1XJY5 T1HZN2 A0A336MRP1 A0A139WPB2 A0A0T6B1P3 A0A1D2NCY7 A0A1W4XK38 A0A1J1JA06 A0A147BKU8 L7MCQ3 A0A182G712 A0A0J7LC56 E2BDJ3 A0A151X9T4 A0A151J3M5 A0A0K8WD88 A0A034WX38 A0A195BEX0 A0A0P6CWD0 A0A0P5SE32 A0A2R7WDR8 A0A162CZK0 A0A0J9U4W3 B4HN86 B4QBR7 Q8T017 A0A0B4LFL6 A0A158NBW6 A0A0A1X3L8 F4W5Q0 A0A0K8U7K7 B4P527 A0A195FLX4 A0A1I8PQJ7 W8ANF3 B3NLK8 E2AKB2 A0A0Q9WEJ6 A0A0A1WRB4 A0A0P4WWE9 A0A1W4V0V5 B4LKT0 B4MYI6 S4RT90 A0A210PTU2 B4J9K2 B4KQU7 A0A226EZ15 R7UET2 A0A1S3KI58 B3MHX5 A0A2C9JVV8 A0A1B0G4D5 Q28XE9 A0A1A9ZNI8 B4GIU8 A0A1A9VIH7 A0A195C9X0 A0A0M3QVF5 A0A3B0J1Q1 V4B1L7 A0A1I8NAB1 A0A1A9XJ55 A0A1A9X3X0 E0VFJ5 A0A2A3ECN6 V9IEC3 A0A088A5Y3 A0A3M7R5K5
Pubmed
26354079
22118469
24438588
25244985
20966253
12364791
+ More
20920257 23761445 28004739 28503490 29209593 18362917 19820115 27289101 29652888 25576852 26483478 20798317 25348373 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24560274 26109357 26109356 21347285 25830018 21719571 17550304 24495485 18057021 28812685 23254933 15562597 15632085 25315136 20566863 30375419
20920257 23761445 28004739 28503490 29209593 18362917 19820115 27289101 29652888 25576852 26483478 20798317 25348373 22936249 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24560274 26109357 26109356 21347285 25830018 21719571 17550304 24495485 18057021 28812685 23254933 15562597 15632085 25315136 20566863 30375419
EMBL
RSAL01000519
RVE41470.1
KQ459606
KPI91033.1
AGBW02010729
OWR47971.1
+ More
NWSH01001385 PCG71454.1 PCG71453.1 ATLV01024097 KE525349 KFB50619.1 AXCM01018056 AXCN02000429 APCN01000165 AAAB01008807 EAA04115.3 DS231885 EDS43190.1 GEBQ01016336 JAT23641.1 GEDC01030438 JAS06860.1 ADMH02001152 ETN63882.1 UFQT01002139 SSX32741.1 GEZM01078793 JAV62620.1 GFAC01005874 JAT93314.1 GFAA01004015 JAT99419.1 ACPB03014434 SSX32740.1 KQ971307 KYB29661.1 LJIG01016219 KRT81258.1 LJIJ01000087 ODN03101.1 CVRI01000075 CRL08366.1 GEGO01003975 JAR91429.1 GACK01003457 JAA61577.1 JXUM01046019 JXUM01046020 KQ561459 KXJ78505.1 LBMM01000004 KMR05388.1 GL447645 EFN86245.1 KQ982373 KYQ57079.1 KQ980286 KYN16927.1 GDHF01003252 JAI49062.1 GAKP01000589 JAC58363.1 KQ976500 KYM83123.1 GDIQ01089288 JAN05449.1 GDIP01140949 JAL62765.1 KK854656 PTY17688.1 LRGB01000024 KZS21669.1 CM002911 KMY94690.1 KMY94691.1 KMY94692.1 CH480816 EDW48368.1 CM000362 EDX07577.1 KMY94689.1 AE013599 AY069629 AAF57785.2 AAL39774.1 AHN56346.1 ADTU01011407 GBXI01008363 JAD05929.1 GL887663 EGI70480.1 GDHF01030004 JAI22310.1 CM000158 EDW91728.2 KQ981491 KYN40989.1 GAMC01019043 GAMC01019041 GAMC01019038 GAMC01019036 JAB87512.1 CH954179 EDV54983.2 GL440211 EFN66125.1 CH940648 KRF80241.1 GBXI01013314 JAD00978.1 GDIP01250715 JAI72686.1 EDW61803.2 KRF80242.1 KRF80243.1 CH963894 EDW77175.2 KRF98591.1 NEDP02005498 OWF39903.1 CH916367 EDW02509.1 CH933808 EDW09296.2 KRG04583.1 LNIX01000001 OXA62779.1 AMQN01001396 KB302197 ELU04586.1 CH902619 EDV36962.2 KPU76549.1 CCAG010007933 CM000071 EAL26367.3 CH479183 EDW35433.1 KQ978068 KYM97510.1 CP012524 ALC42329.1 OUUW01000001 SPP74917.1 KB200521 ESP01211.1 DS235114 EEB12151.1 KZ288284 PBC29505.1 JR040174 AEY59012.1 REGN01004166 RNA18826.1
NWSH01001385 PCG71454.1 PCG71453.1 ATLV01024097 KE525349 KFB50619.1 AXCM01018056 AXCN02000429 APCN01000165 AAAB01008807 EAA04115.3 DS231885 EDS43190.1 GEBQ01016336 JAT23641.1 GEDC01030438 JAS06860.1 ADMH02001152 ETN63882.1 UFQT01002139 SSX32741.1 GEZM01078793 JAV62620.1 GFAC01005874 JAT93314.1 GFAA01004015 JAT99419.1 ACPB03014434 SSX32740.1 KQ971307 KYB29661.1 LJIG01016219 KRT81258.1 LJIJ01000087 ODN03101.1 CVRI01000075 CRL08366.1 GEGO01003975 JAR91429.1 GACK01003457 JAA61577.1 JXUM01046019 JXUM01046020 KQ561459 KXJ78505.1 LBMM01000004 KMR05388.1 GL447645 EFN86245.1 KQ982373 KYQ57079.1 KQ980286 KYN16927.1 GDHF01003252 JAI49062.1 GAKP01000589 JAC58363.1 KQ976500 KYM83123.1 GDIQ01089288 JAN05449.1 GDIP01140949 JAL62765.1 KK854656 PTY17688.1 LRGB01000024 KZS21669.1 CM002911 KMY94690.1 KMY94691.1 KMY94692.1 CH480816 EDW48368.1 CM000362 EDX07577.1 KMY94689.1 AE013599 AY069629 AAF57785.2 AAL39774.1 AHN56346.1 ADTU01011407 GBXI01008363 JAD05929.1 GL887663 EGI70480.1 GDHF01030004 JAI22310.1 CM000158 EDW91728.2 KQ981491 KYN40989.1 GAMC01019043 GAMC01019041 GAMC01019038 GAMC01019036 JAB87512.1 CH954179 EDV54983.2 GL440211 EFN66125.1 CH940648 KRF80241.1 GBXI01013314 JAD00978.1 GDIP01250715 JAI72686.1 EDW61803.2 KRF80242.1 KRF80243.1 CH963894 EDW77175.2 KRF98591.1 NEDP02005498 OWF39903.1 CH916367 EDW02509.1 CH933808 EDW09296.2 KRG04583.1 LNIX01000001 OXA62779.1 AMQN01001396 KB302197 ELU04586.1 CH902619 EDV36962.2 KPU76549.1 CCAG010007933 CM000071 EAL26367.3 CH479183 EDW35433.1 KQ978068 KYM97510.1 CP012524 ALC42329.1 OUUW01000001 SPP74917.1 KB200521 ESP01211.1 DS235114 EEB12151.1 KZ288284 PBC29505.1 JR040174 AEY59012.1 REGN01004166 RNA18826.1
Proteomes
UP000283053
UP000053268
UP000007151
UP000218220
UP000075900
UP000030765
+ More
UP000069272 UP000075883 UP000075884 UP000076408 UP000075901 UP000075885 UP000075920 UP000075886 UP000075840 UP000075882 UP000076407 UP000075903 UP000075902 UP000007062 UP000002320 UP000000673 UP000015103 UP000007266 UP000094527 UP000192223 UP000183832 UP000069940 UP000249989 UP000036403 UP000008237 UP000075809 UP000078492 UP000078540 UP000076858 UP000001292 UP000000304 UP000000803 UP000005205 UP000007755 UP000002282 UP000078541 UP000095300 UP000008711 UP000000311 UP000008792 UP000192221 UP000007798 UP000245300 UP000242188 UP000001070 UP000009192 UP000198287 UP000014760 UP000085678 UP000007801 UP000076420 UP000092444 UP000001819 UP000092445 UP000008744 UP000078200 UP000078542 UP000092553 UP000268350 UP000030746 UP000095301 UP000092443 UP000091820 UP000009046 UP000242457 UP000005203 UP000276133
UP000069272 UP000075883 UP000075884 UP000076408 UP000075901 UP000075885 UP000075920 UP000075886 UP000075840 UP000075882 UP000076407 UP000075903 UP000075902 UP000007062 UP000002320 UP000000673 UP000015103 UP000007266 UP000094527 UP000192223 UP000183832 UP000069940 UP000249989 UP000036403 UP000008237 UP000075809 UP000078492 UP000078540 UP000076858 UP000001292 UP000000304 UP000000803 UP000005205 UP000007755 UP000002282 UP000078541 UP000095300 UP000008711 UP000000311 UP000008792 UP000192221 UP000007798 UP000245300 UP000242188 UP000001070 UP000009192 UP000198287 UP000014760 UP000085678 UP000007801 UP000076420 UP000092444 UP000001819 UP000092445 UP000008744 UP000078200 UP000078542 UP000092553 UP000268350 UP000030746 UP000095301 UP000092443 UP000091820 UP000009046 UP000242457 UP000005203 UP000276133
PRIDE
Pfam
PF00615 RGS
SUPFAM
SSF48097
SSF48097
Gene 3D
ProteinModelPortal
A0A3S2TBZ1
A0A194PDE7
A0A212F2M3
A0A2A4JIQ1
A0A2A4JJ59
A0A182RP65
+ More
A0A084WK75 A0A182FC83 A0A182M6Q9 A0A182NK54 A0A182YEL5 A0A182T0R3 A0A182P0N8 A0A182WLI8 A0A182QDH1 A0A1D6T4Q2 A0A182KQE1 A0A182WWZ6 A0A182V695 A0A182I2J3 A0A182TH05 Q7QIY2 B0WC38 A0A1B6LIX0 A0A1B6C0T6 W5JHN4 A0A336MT64 A0A1Y1KR19 A0A1E1X245 A0A1E1XJY5 T1HZN2 A0A336MRP1 A0A139WPB2 A0A0T6B1P3 A0A1D2NCY7 A0A1W4XK38 A0A1J1JA06 A0A147BKU8 L7MCQ3 A0A182G712 A0A0J7LC56 E2BDJ3 A0A151X9T4 A0A151J3M5 A0A0K8WD88 A0A034WX38 A0A195BEX0 A0A0P6CWD0 A0A0P5SE32 A0A2R7WDR8 A0A162CZK0 A0A0J9U4W3 B4HN86 B4QBR7 Q8T017 A0A0B4LFL6 A0A158NBW6 A0A0A1X3L8 F4W5Q0 A0A0K8U7K7 B4P527 A0A195FLX4 A0A1I8PQJ7 W8ANF3 B3NLK8 E2AKB2 A0A0Q9WEJ6 A0A0A1WRB4 A0A0P4WWE9 A0A1W4V0V5 B4LKT0 B4MYI6 S4RT90 A0A210PTU2 B4J9K2 B4KQU7 A0A226EZ15 R7UET2 A0A1S3KI58 B3MHX5 A0A2C9JVV8 A0A1B0G4D5 Q28XE9 A0A1A9ZNI8 B4GIU8 A0A1A9VIH7 A0A195C9X0 A0A0M3QVF5 A0A3B0J1Q1 V4B1L7 A0A1I8NAB1 A0A1A9XJ55 A0A1A9X3X0 E0VFJ5 A0A2A3ECN6 V9IEC3 A0A088A5Y3 A0A3M7R5K5
A0A084WK75 A0A182FC83 A0A182M6Q9 A0A182NK54 A0A182YEL5 A0A182T0R3 A0A182P0N8 A0A182WLI8 A0A182QDH1 A0A1D6T4Q2 A0A182KQE1 A0A182WWZ6 A0A182V695 A0A182I2J3 A0A182TH05 Q7QIY2 B0WC38 A0A1B6LIX0 A0A1B6C0T6 W5JHN4 A0A336MT64 A0A1Y1KR19 A0A1E1X245 A0A1E1XJY5 T1HZN2 A0A336MRP1 A0A139WPB2 A0A0T6B1P3 A0A1D2NCY7 A0A1W4XK38 A0A1J1JA06 A0A147BKU8 L7MCQ3 A0A182G712 A0A0J7LC56 E2BDJ3 A0A151X9T4 A0A151J3M5 A0A0K8WD88 A0A034WX38 A0A195BEX0 A0A0P6CWD0 A0A0P5SE32 A0A2R7WDR8 A0A162CZK0 A0A0J9U4W3 B4HN86 B4QBR7 Q8T017 A0A0B4LFL6 A0A158NBW6 A0A0A1X3L8 F4W5Q0 A0A0K8U7K7 B4P527 A0A195FLX4 A0A1I8PQJ7 W8ANF3 B3NLK8 E2AKB2 A0A0Q9WEJ6 A0A0A1WRB4 A0A0P4WWE9 A0A1W4V0V5 B4LKT0 B4MYI6 S4RT90 A0A210PTU2 B4J9K2 B4KQU7 A0A226EZ15 R7UET2 A0A1S3KI58 B3MHX5 A0A2C9JVV8 A0A1B0G4D5 Q28XE9 A0A1A9ZNI8 B4GIU8 A0A1A9VIH7 A0A195C9X0 A0A0M3QVF5 A0A3B0J1Q1 V4B1L7 A0A1I8NAB1 A0A1A9XJ55 A0A1A9X3X0 E0VFJ5 A0A2A3ECN6 V9IEC3 A0A088A5Y3 A0A3M7R5K5
PDB
4IGU
E-value=2.29856e-54,
Score=532
Ontologies
KEGG
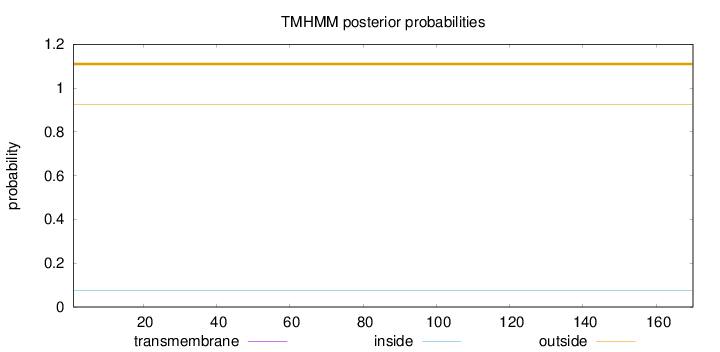
Topology
Length:
170
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07609
outside
1 - 170
Population Genetic Test Statistics
Pi
206.620259
Theta
173.826964
Tajima's D
1.354773
CLR
0
CSRT
0.750612469376531
Interpretation
Uncertain
