Gene
KWMTBOMO10652
Annotation
reverse_transcriptase_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.397
Sequence
CDS
ATGAAGCTCGGTCGCGCTCCCGATTCCGTTTCCGTCACGAGGACTGTGGTGGATTGGCACACGCTGGGCATCAGCCTGGCTGAATCTGATCCACCATCGCTCCCGTTTAGCCCGGACTCTACGCCTCTACGTCTACGTCTCCTCAGGATACCGCTGAAGCCACACATCACCTCGACATTAGATAGGTCATCGAAACAAGTTGTAGCGGAGGACTTCCTTCACCGCTTCAAATTGTCCGACGATATTAGGGAACTCCTTAGAGCTAAGAACGCCTCGATACGCGCCTACGACAGGTATCCTACTGCGGAAAATCGTATTCGAATGCGTGTCCTACAACGCGACGTAAAGTCTCGCATCGCCGAAGTCCGAGATGTCAGATGGTCTGATTTCTTAGAAGGACTCGCGCCCTCCCAAAGCTCTTACTACCGCTTAGCTCGTACTCTCAAATCGGATACGGTAGTAACTACGCCCCCCCTCGCAGGCCCCTCAGGTCGACTCGCGGCGTTCGATGATGACGAAAAAGCAGAGCCACTGGCCGATACATTGCAAACCCAGTGCAAGCCCAGCACTCAATCCGTGGACCCTGTTCATGTAGAATTAGTAGACAGTGAGGTAGAACGCAGAGCCTGCTTGCCACCCTCGGATGCGTTACCACCCGTCACCCCGATGGAAGTTAAAGACTTGATCAAAGACCTACGTCCTCGCAAGGCTCCCGGTTCCGACGGACTATCCAACCGCGTTATTAAACTTCTACCCGTCCAACTCATCGTGATGTTGGCATCTATTTTCAATGCCGCTATGGCGAACTGTATCTTTCCCGCGGTGTGGAAAGAAGCGGACGTTATCGGCATACATAAACCCGGTAAACCAAAAAATCATCCGACGAGCTACCGCCCGATTAGCCTCCTCATGTCTCTAGGCAAACTGTATGAGCGTCTGCTCTACAAACGCCTGAGAGACTTCGTCTCATCCAAGGGCAGTCTCATCGATGAACAATTCGGATTCCGTACAAATCACTCATGCGTTCAACAGGTGCACCGCCTCACGGAGCACATTCTTGTGGGGCTTAATCGACCAAAACCGCGATACACGGGAGCTCTCTTTTTCGACGTCGAAGCGTTCGACAAAGTCTGGCACAACGGTTTGATTTTGAAACTATTCAACATGGGCGTGCCGGATAGTCTCGTGCTCACCATACGGGACTTCTTGTCGAACCGCTCTTTTCGATATCGAGTCGAGGGAACCTGCTCCTCCCCACGATCTCTCACAGCTGGAGTCCCGCAAGGCTCTGTCCTCTCACCCCTCCTATTTAGCTTATTCGTCAACGATATTCCCCGGTCGCCGCCGACCCAGTTAGCCTTATTCGCCGACAACACGACTGTTTACTATTCCAGTAGAAACAAGTCCCTAATCGCGAAGAAGCTTCAGAGCGCAGCCCTAGCCCTAGGACAGTGGTTCCGAAAATGGCGCATAGACATCAACCCAGCGAAAAGTACTGCGGTGCTATTTCAGAGGGGAAGCTCCACACGGATTTCCTCCCGGATTAGGAGGAGGAATCTCACACCCCCGATTACTCTCTTTAGACAACCCATACCCTGGGCCGCTGACTACTCCCCGAATCCTGATCACGCAGGAGCCAGTCACCGTCGACGCCCTAGACACGTCCTTACGGATCCATCAGATCCAATAACCTTTGCATTAGACGCCTTCAGCTTTAACTCTAGGAGCAGGCTTAGGGACCACGATAACCGTACTCGTCGAAATCGACAAAGAGTCAATGTGCAACCTAAGCCATGCATCAGCCCGCTGAGTTTCTCGCCGGGTTTTCTCAGCGGGTCGCGATTCCGATCCGGTAGTAAATTCATTCGCGAAGCAGCTACTCTTGAGCTGTTAGGTCTCCTTCGGAAGCGCTCGGGCAGCTATTAG
Protein
MKLGRAPDSVSVTRTVVDWHTLGISLAESDPPSLPFSPDSTPLRLRLLRIPLKPHITSTLDRSSKQVVAEDFLHRFKLSDDIRELLRAKNASIRAYDRYPTAENRIRMRVLQRDVKSRIAEVRDVRWSDFLEGLAPSQSSYYRLARTLKSDTVVTTPPLAGPSGRLAAFDDDEKAEPLADTLQTQCKPSTQSVDPVHVELVDSEVERRACLPPSDALPPVTPMEVKDLIKDLRPRKAPGSDGLSNRVIKLLPVQLIVMLASIFNAAMANCIFPAVWKEADVIGIHKPGKPKNHPTSYRPISLLMSLGKLYERLLYKRLRDFVSSKGSLIDEQFGFRTNHSCVQQVHRLTEHILVGLNRPKPRYTGALFFDVEAFDKVWHNGLILKLFNMGVPDSLVLTIRDFLSNRSFRYRVEGTCSSPRSLTAGVPQGSVLSPLLFSLFVNDIPRSPPTQLALFADNTTVYYSSRNKSLIAKKLQSAALALGQWFRKWRIDINPAKSTAVLFQRGSSTRISSRIRRRNLTPPITLFRQPIPWAADYSPNPDHAGASHRRRPRHVLTDPSDPITFALDAFSFNSRSRLRDHDNRTRRNRQRVNVQPKPCISPLSFSPGFLSGSRFRSGSKFIREAATLELLGLLRKRSGSY
Summary
Uniprot
EMBL
U07847
AAA17752.1
AB018558
BAA76304.1
AB055391
BAB21761.1
+ More
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 GGMS01006359 MBY75562.1 GGMS01017914 MBY87117.1 ABLF02042518 ABLF02057746 ABLF02040174 ABLF02040183 ABLF02055266 ABLF02028535 ABLF02028545 ABLF02024246 GGMR01013409 MBY26028.1 ABLF02008463 ABLF02008153 ABLF02008462 ABLF02008464
KQ459265 KPJ02064.1 AF081103 AAC72921.1 GEZM01008457 JAV94772.1 GGMS01006359 MBY75562.1 GGMS01017914 MBY87117.1 ABLF02042518 ABLF02057746 ABLF02040174 ABLF02040183 ABLF02055266 ABLF02028535 ABLF02028545 ABLF02024246 GGMR01013409 MBY26028.1 ABLF02008463 ABLF02008153 ABLF02008462 ABLF02008464
Proteomes
Interpro
SUPFAM
SSF56219
SSF56219
Gene 3D
ProteinModelPortal
Ontologies
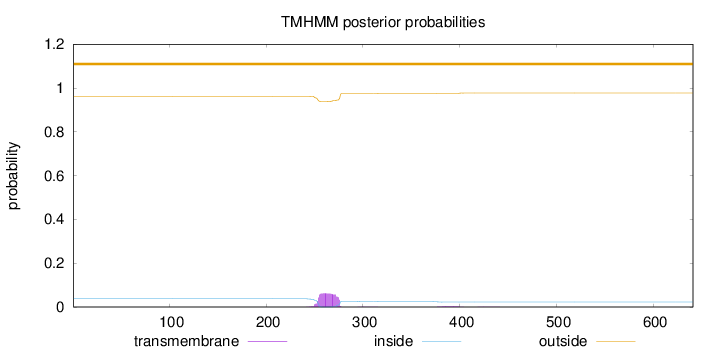
Topology
Length:
641
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.45911000000001
Exp number, first 60 AAs:
9e-05
Total prob of N-in:
0.03778
outside
1 - 641
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0.091468
CSRT
0
Interpretation
Uncertain
