Gene
KWMTBOMO10641
Pre Gene Modal
BGIBMGA003964
Annotation
hypothetical_protein_KGM_04995_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 4.93
Sequence
CDS
ATGATTCCAAGACGCAAACACAACGCATCCTCGGGCTCCCTGCCGCCTCCCGACGACAAGAACGCCAGCAATGACTACATCCTGTCGGAGGGAACGTCGTGCGAGCGGCTATGCGCTGCGCTGCGGCGTCGGTTGAACCGCAAGACGCTGCACAAGCGCGTCCCGGTGACGGCGTGGCTGCCGCAGTACAACGCGGAGAAGGCGATCGGCGACCTGATCGCCGGCATCACGGTGGGCCTCACGGTCATCCCGCAGTCGCTAGCGTACTCCAACATCGCGGGCCTGCCGCCGCAGCACGGCCTGTACGGCTCGTTCTTGGGCTGCTTCGTGTACATCGTGCTGGGCGGGTGTCGCGCCGTGCCCGCCGGCCCCACCGCCATCGCCTCGCTCTTGACTTGGCAGGTCGCCGGCGGTGTGGTCGAGAAGGCCATTCTGCTGACGCTCCTGGCGGGAATAGTCGAGCTCATGATGGGTGTCCTCGGGCTCGGCTTTTTGATTAACTTCGTGTCGGGTCCGGTCTCGTCGGGGTTCACGTCGGCCGTGGCACTGATGATCGCTACGTCGCAGGTCAAGGACATGTTCGCAATTTCAGTCACCGGTGAAACCTTCCTCCAGCAGTGGATCTCAATATTCCGAAACCTGCACGCTGCTGCACTGTGGGACCCCGTCCTCGGCTTCGCCTGCATCGCCTTGCTCCTGCTCTTGAGGAAAATTGGAATGATAAAGCTAGGTGGCAATTCTGAAAACGGACCGAACACTCGACAGAAAGTGGTGACGAAGTGCTTGTGGCTGCTGGGGACCTGTCGGAACGCGATCGTGGTGGTGGCCACCGGCGTGCTCGGCTACTGGTTCGTCGCCACCCACGGCTCCTCGCCCGTCCGCCTCATGGGTGCGATCCCGTCCGGCGTGCCCCACCCGCAGCTGCCGCCGTTCAGCTACGTGCGCGCCGACAACACGACCGCCGACTTCCTGGACATGGTGTCGGAGCTGGGCTCGGGGCTGCTGGTCATCCCGCTCATCGTGCTGCTGGAGGACATCGCCATCTGCAAGGCGTTCTCTGACGGGCGCACCATCGACGCGACGCAGGAGATGATCGCGCTGGGCGTGGCCAGCATCGCCAACTCGTTCGTGCAGGCGTACCCGGGCGGCGGCTCGCTGGCTCGCTCCGTCGTCTGCAACGGCTCTGGCGTCAAGACCACCTTCAACGGACTCTACACCGGAATTATGGTGATCCTTGCCCTGCAGTTCTTCACGCAGTACTTTGAGTATATCCCGAAGGCCGCTCTCGCTGCCGTCATCATAACTGCGATCCTGTTTATGGTGGAATACAACGTGATCAAGCCCATGTGGAGAGCTAAAAAACTGGATCTCATCCCGGGCGTGGGCACGTTCGTGCTGTGCCTGACGCTACCGATCGAGCTGGGCATCCTGACGGGCGTGATCGTCAACATCATCTTCATCCTGTACCACGCCGCTCGGCCCAAGTTCTCGGTGGAGATGTTGAAGACGGAGCAGGGCGTGGAGTACCTCATGGTGACCCCGGACCGCTGCCTCATGTTCCCCTCGGTGGACTACGTGCGGCGCCTGGTGACCAAGTTCGCGGGCAGCGGCTCCGTGCCCGTGGTGCTGGAGTGCACGCACATCTACAGCGCGGACTACACCGCCGCCAAGAGCATCGAGCAGCTCACCGCAGACTTCCACGCGCGACGGCAGCCGCTCTACTTCTACAACGTCAAGCCTTCCGTCTGCTCGATCTTCGAGGCGGTCGCGAAGCCCGAGCACTTCGTGGTGTTCTACGAGGACGAGGAGCTGGACCGGCTGCTCGCGGCGGACGAGCGCCTCGCGCCCACCAAGCAGGCGCCTCTCGACGCCTGA
Protein
MIPRRKHNASSGSLPPPDDKNASNDYILSEGTSCERLCAALRRRLNRKTLHKRVPVTAWLPQYNAEKAIGDLIAGITVGLTVIPQSLAYSNIAGLPPQHGLYGSFLGCFVYIVLGGCRAVPAGPTAIASLLTWQVAGGVVEKAILLTLLAGIVELMMGVLGLGFLINFVSGPVSSGFTSAVALMIATSQVKDMFAISVTGETFLQQWISIFRNLHAAALWDPVLGFACIALLLLLRKIGMIKLGGNSENGPNTRQKVVTKCLWLLGTCRNAIVVVATGVLGYWFVATHGSSPVRLMGAIPSGVPHPQLPPFSYVRADNTTADFLDMVSELGSGLLVIPLIVLLEDIAICKAFSDGRTIDATQEMIALGVASIANSFVQAYPGGGSLARSVVCNGSGVKTTFNGLYTGIMVILALQFFTQYFEYIPKAALAAVIITAILFMVEYNVIKPMWRAKKLDLIPGVGTFVLCLTLPIELGILTGVIVNIIFILYHAARPKFSVEMLKTEQGVEYLMVTPDRCLMFPSVDYVRRLVTKFAGSGSVPVVLECTHIYSADYTAAKSIEQLTADFHARRQPLYFYNVKPSVCSIFEAVAKPEHFVVFYEDEELDRLLAADERLAPTKQAPLDA
Summary
Similarity
Belongs to the SLC26A/SulP transporter (TC 2.A.53) family.
Uniprot
H9J377
A0A212F946
A0A2H1W269
A0A194QDN0
A0A2W1BHD9
A0A2A4JI43
+ More
A0A3S2NCS5 S4NP74 A0A0C9QYS7 E2BS57 A0A3L8D3E8 A0A310SI74 A0A1B0DR61 A0A154PPK6 D6WUN9 A0A2A3EB20 A0A1B0CLE2 A0A026W8W4 A0A1L8DEP9 V9IDC5 A0A088A7G0 A0A182IXP3 E9ILM7 A0A182RY98 A0A182F6K0 A0A182M4V4 A0A2M4A5J3 A0A1Q3FN70 A0A2M3ZFE6 A0A2M4BGE8 A0A2M4BGI5 W5JUZ8 A0A195CH18 A0A182QWG3 A0A182W2Q5 A0A0M8ZSJ6 A0A182XVH8 A0A182NGD0 A0A1W4XLX0 Q17AX2 A0A195EMB3 F4WFB0 B0WDI9 A0A0K8TQT4 A0A182G269 A0A182GXB2 A0A195BYN7 U5EHL8 A0A151XF28 A0A0L0C6S2 A0A023EVP6 A0A2P8XXV8 A0A336MYI8 A0A336L6S7 A0A1L8DDY7 A0A336MYE0 A0A067RKF7 A0A336N4K8 A0A1S4FDF4 Q176H5 Q7QBV8 A0A1Q3FHN8 A0A1Q3FH17 A0A182HTD9 A0A182LFI7 A0A182P863 B0WZD9 W8B4W6 A0A182HF16 A0A0A1XJ05 A0A0A1XCP3 N6TDV8 A0A182U8Y5 U4UHE5 A0A182MYK8 A0A182UME3 Q9VC29 B4PV34 Q29BI6 A0A182JU99 B4QTT8 A0A182FU53 A0A0K8UCR2 A0A336L4K1 W5J1Z2 A0A3B0JIL0 A0A182QFM7 A0A1B0DRA4 B3LV12 A0A182XEJ3 Q9U6Q2 A0A182UAG7 Q16ED9 A0A084WHU1 A0A182UN25 Q7PV84 A0A182HQ40 A0A182XNB9 A0A1B6IYY2 A0A182R5D4
A0A3S2NCS5 S4NP74 A0A0C9QYS7 E2BS57 A0A3L8D3E8 A0A310SI74 A0A1B0DR61 A0A154PPK6 D6WUN9 A0A2A3EB20 A0A1B0CLE2 A0A026W8W4 A0A1L8DEP9 V9IDC5 A0A088A7G0 A0A182IXP3 E9ILM7 A0A182RY98 A0A182F6K0 A0A182M4V4 A0A2M4A5J3 A0A1Q3FN70 A0A2M3ZFE6 A0A2M4BGE8 A0A2M4BGI5 W5JUZ8 A0A195CH18 A0A182QWG3 A0A182W2Q5 A0A0M8ZSJ6 A0A182XVH8 A0A182NGD0 A0A1W4XLX0 Q17AX2 A0A195EMB3 F4WFB0 B0WDI9 A0A0K8TQT4 A0A182G269 A0A182GXB2 A0A195BYN7 U5EHL8 A0A151XF28 A0A0L0C6S2 A0A023EVP6 A0A2P8XXV8 A0A336MYI8 A0A336L6S7 A0A1L8DDY7 A0A336MYE0 A0A067RKF7 A0A336N4K8 A0A1S4FDF4 Q176H5 Q7QBV8 A0A1Q3FHN8 A0A1Q3FH17 A0A182HTD9 A0A182LFI7 A0A182P863 B0WZD9 W8B4W6 A0A182HF16 A0A0A1XJ05 A0A0A1XCP3 N6TDV8 A0A182U8Y5 U4UHE5 A0A182MYK8 A0A182UME3 Q9VC29 B4PV34 Q29BI6 A0A182JU99 B4QTT8 A0A182FU53 A0A0K8UCR2 A0A336L4K1 W5J1Z2 A0A3B0JIL0 A0A182QFM7 A0A1B0DRA4 B3LV12 A0A182XEJ3 Q9U6Q2 A0A182UAG7 Q16ED9 A0A084WHU1 A0A182UN25 Q7PV84 A0A182HQ40 A0A182XNB9 A0A1B6IYY2 A0A182R5D4
Pubmed
19121390
22118469
26354079
28756777
23622113
20798317
+ More
30249741 18362917 19820115 24508170 21282665 20920257 23761445 25244985 17510324 21719571 26369729 26483478 26108605 24945155 29403074 24845553 12364791 14747013 17210077 20966253 24495485 25830018 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 15632085 24438588
30249741 18362917 19820115 24508170 21282665 20920257 23761445 25244985 17510324 21719571 26369729 26483478 26108605 24945155 29403074 24845553 12364791 14747013 17210077 20966253 24495485 25830018 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 17550304 15632085 24438588
EMBL
BABH01036732
AGBW02009651
OWR50251.1
ODYU01005834
SOQ47113.1
KQ459144
+ More
KPJ03658.1 KZ150383 PZC71083.1 NWSH01001388 PCG71446.1 RSAL01000214 RVE44146.1 GAIX01011959 JAA80601.1 GBYB01000869 GBYB01000870 GBYB01000871 JAG70636.1 JAG70637.1 JAG70638.1 GL450151 EFN81449.1 QOIP01000014 RLU15025.1 KQ759878 OAD62248.1 AJVK01019712 KQ435012 KZC13813.1 KQ971363 EFA07819.1 KZ288296 PBC28927.1 AJWK01017158 AJWK01017159 AJWK01017160 AJWK01017161 AJWK01017162 AJWK01017163 KK107389 EZA51469.1 GFDF01009163 JAV04921.1 JR039342 AEY58647.1 GL764129 EFZ18364.1 AXCM01000024 GGFK01002679 MBW36000.1 GFDL01006048 JAV28997.1 GGFM01006501 MBW27252.1 GGFJ01002995 MBW52136.1 GGFJ01002996 MBW52137.1 ADMH02000060 ETN67956.1 KQ977791 KYM99726.1 AXCN02001614 KQ435918 KOX68743.1 CH477329 EAT43399.1 KQ978691 KYN29266.1 GL888115 EGI67161.1 DS231898 EDS44598.1 GDAI01000881 JAI16722.1 JXUM01138468 KQ568748 KXJ68894.1 JXUM01095035 KQ564168 KXJ72645.1 KQ976396 KYM93046.1 GANO01002964 JAB56907.1 KQ982215 KYQ58931.1 JRES01000835 KNC27932.1 GAPW01000467 JAC13131.1 PYGN01001181 PSN36843.1 UFQT01001527 SSX30928.1 UFQS01001800 UFQT01001800 SSX12373.1 SSX31824.1 GFDF01009418 JAV04666.1 UFQS01002399 UFQT01002399 SSX13948.1 SSX33367.1 KK852421 KDR24302.1 SSX13949.1 SSX33368.1 CH477387 EAT42041.1 AAAB01008859 EAA07491.5 GFDL01007964 JAV27081.1 GFDL01008197 JAV26848.1 APCN01000481 DS232206 EDS37453.1 GAMC01018214 GAMC01018213 JAB88341.1 JXUM01036390 JXUM01036391 JXUM01036392 JXUM01036393 JXUM01036394 KQ561093 KXJ79726.1 GBXI01003774 JAD10518.1 GBXI01005570 JAD08722.1 APGK01041487 APGK01041488 APGK01041489 APGK01041490 KB740994 ENN75938.1 KB632356 ERL93394.1 AE014297 AY071258 AAF56347.1 AAL48880.1 ADV37372.1 CM000160 EDW98883.1 KRK04512.1 CM000070 EAL27012.2 CM000364 EDX14292.1 GDHF01028879 GDHF01028169 GDHF01003223 JAI23435.1 JAI24145.1 JAI49091.1 SSX12374.1 SSX31825.1 ADMH02002215 ETN57756.1 OUUW01000005 SPP80152.1 AXCN02001041 AJVK01019779 CH902617 EDV42484.1 AF180728 AAD53951.1 CH478830 EAT32596.1 ATLV01023888 KE525347 KFB49785.1 AAAB01008986 EAA00286.5 APCN01003291 GECU01015548 GECU01001209 JAS92158.1 JAT06498.1
KPJ03658.1 KZ150383 PZC71083.1 NWSH01001388 PCG71446.1 RSAL01000214 RVE44146.1 GAIX01011959 JAA80601.1 GBYB01000869 GBYB01000870 GBYB01000871 JAG70636.1 JAG70637.1 JAG70638.1 GL450151 EFN81449.1 QOIP01000014 RLU15025.1 KQ759878 OAD62248.1 AJVK01019712 KQ435012 KZC13813.1 KQ971363 EFA07819.1 KZ288296 PBC28927.1 AJWK01017158 AJWK01017159 AJWK01017160 AJWK01017161 AJWK01017162 AJWK01017163 KK107389 EZA51469.1 GFDF01009163 JAV04921.1 JR039342 AEY58647.1 GL764129 EFZ18364.1 AXCM01000024 GGFK01002679 MBW36000.1 GFDL01006048 JAV28997.1 GGFM01006501 MBW27252.1 GGFJ01002995 MBW52136.1 GGFJ01002996 MBW52137.1 ADMH02000060 ETN67956.1 KQ977791 KYM99726.1 AXCN02001614 KQ435918 KOX68743.1 CH477329 EAT43399.1 KQ978691 KYN29266.1 GL888115 EGI67161.1 DS231898 EDS44598.1 GDAI01000881 JAI16722.1 JXUM01138468 KQ568748 KXJ68894.1 JXUM01095035 KQ564168 KXJ72645.1 KQ976396 KYM93046.1 GANO01002964 JAB56907.1 KQ982215 KYQ58931.1 JRES01000835 KNC27932.1 GAPW01000467 JAC13131.1 PYGN01001181 PSN36843.1 UFQT01001527 SSX30928.1 UFQS01001800 UFQT01001800 SSX12373.1 SSX31824.1 GFDF01009418 JAV04666.1 UFQS01002399 UFQT01002399 SSX13948.1 SSX33367.1 KK852421 KDR24302.1 SSX13949.1 SSX33368.1 CH477387 EAT42041.1 AAAB01008859 EAA07491.5 GFDL01007964 JAV27081.1 GFDL01008197 JAV26848.1 APCN01000481 DS232206 EDS37453.1 GAMC01018214 GAMC01018213 JAB88341.1 JXUM01036390 JXUM01036391 JXUM01036392 JXUM01036393 JXUM01036394 KQ561093 KXJ79726.1 GBXI01003774 JAD10518.1 GBXI01005570 JAD08722.1 APGK01041487 APGK01041488 APGK01041489 APGK01041490 KB740994 ENN75938.1 KB632356 ERL93394.1 AE014297 AY071258 AAF56347.1 AAL48880.1 ADV37372.1 CM000160 EDW98883.1 KRK04512.1 CM000070 EAL27012.2 CM000364 EDX14292.1 GDHF01028879 GDHF01028169 GDHF01003223 JAI23435.1 JAI24145.1 JAI49091.1 SSX12374.1 SSX31825.1 ADMH02002215 ETN57756.1 OUUW01000005 SPP80152.1 AXCN02001041 AJVK01019779 CH902617 EDV42484.1 AF180728 AAD53951.1 CH478830 EAT32596.1 ATLV01023888 KE525347 KFB49785.1 AAAB01008986 EAA00286.5 APCN01003291 GECU01015548 GECU01001209 JAS92158.1 JAT06498.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000283053
UP000008237
+ More
UP000279307 UP000092462 UP000076502 UP000007266 UP000242457 UP000092461 UP000053097 UP000005203 UP000075880 UP000075900 UP000069272 UP000075883 UP000000673 UP000078542 UP000075886 UP000075920 UP000053105 UP000076408 UP000075884 UP000192223 UP000008820 UP000078492 UP000007755 UP000002320 UP000069940 UP000249989 UP000078540 UP000075809 UP000037069 UP000245037 UP000027135 UP000007062 UP000075840 UP000075882 UP000075885 UP000019118 UP000075902 UP000030742 UP000075903 UP000000803 UP000002282 UP000001819 UP000075881 UP000000304 UP000268350 UP000007801 UP000076407 UP000030765
UP000279307 UP000092462 UP000076502 UP000007266 UP000242457 UP000092461 UP000053097 UP000005203 UP000075880 UP000075900 UP000069272 UP000075883 UP000000673 UP000078542 UP000075886 UP000075920 UP000053105 UP000076408 UP000075884 UP000192223 UP000008820 UP000078492 UP000007755 UP000002320 UP000069940 UP000249989 UP000078540 UP000075809 UP000037069 UP000245037 UP000027135 UP000007062 UP000075840 UP000075882 UP000075885 UP000019118 UP000075902 UP000030742 UP000075903 UP000000803 UP000002282 UP000001819 UP000075881 UP000000304 UP000268350 UP000007801 UP000076407 UP000030765
Interpro
Gene 3D
ProteinModelPortal
H9J377
A0A212F946
A0A2H1W269
A0A194QDN0
A0A2W1BHD9
A0A2A4JI43
+ More
A0A3S2NCS5 S4NP74 A0A0C9QYS7 E2BS57 A0A3L8D3E8 A0A310SI74 A0A1B0DR61 A0A154PPK6 D6WUN9 A0A2A3EB20 A0A1B0CLE2 A0A026W8W4 A0A1L8DEP9 V9IDC5 A0A088A7G0 A0A182IXP3 E9ILM7 A0A182RY98 A0A182F6K0 A0A182M4V4 A0A2M4A5J3 A0A1Q3FN70 A0A2M3ZFE6 A0A2M4BGE8 A0A2M4BGI5 W5JUZ8 A0A195CH18 A0A182QWG3 A0A182W2Q5 A0A0M8ZSJ6 A0A182XVH8 A0A182NGD0 A0A1W4XLX0 Q17AX2 A0A195EMB3 F4WFB0 B0WDI9 A0A0K8TQT4 A0A182G269 A0A182GXB2 A0A195BYN7 U5EHL8 A0A151XF28 A0A0L0C6S2 A0A023EVP6 A0A2P8XXV8 A0A336MYI8 A0A336L6S7 A0A1L8DDY7 A0A336MYE0 A0A067RKF7 A0A336N4K8 A0A1S4FDF4 Q176H5 Q7QBV8 A0A1Q3FHN8 A0A1Q3FH17 A0A182HTD9 A0A182LFI7 A0A182P863 B0WZD9 W8B4W6 A0A182HF16 A0A0A1XJ05 A0A0A1XCP3 N6TDV8 A0A182U8Y5 U4UHE5 A0A182MYK8 A0A182UME3 Q9VC29 B4PV34 Q29BI6 A0A182JU99 B4QTT8 A0A182FU53 A0A0K8UCR2 A0A336L4K1 W5J1Z2 A0A3B0JIL0 A0A182QFM7 A0A1B0DRA4 B3LV12 A0A182XEJ3 Q9U6Q2 A0A182UAG7 Q16ED9 A0A084WHU1 A0A182UN25 Q7PV84 A0A182HQ40 A0A182XNB9 A0A1B6IYY2 A0A182R5D4
A0A3S2NCS5 S4NP74 A0A0C9QYS7 E2BS57 A0A3L8D3E8 A0A310SI74 A0A1B0DR61 A0A154PPK6 D6WUN9 A0A2A3EB20 A0A1B0CLE2 A0A026W8W4 A0A1L8DEP9 V9IDC5 A0A088A7G0 A0A182IXP3 E9ILM7 A0A182RY98 A0A182F6K0 A0A182M4V4 A0A2M4A5J3 A0A1Q3FN70 A0A2M3ZFE6 A0A2M4BGE8 A0A2M4BGI5 W5JUZ8 A0A195CH18 A0A182QWG3 A0A182W2Q5 A0A0M8ZSJ6 A0A182XVH8 A0A182NGD0 A0A1W4XLX0 Q17AX2 A0A195EMB3 F4WFB0 B0WDI9 A0A0K8TQT4 A0A182G269 A0A182GXB2 A0A195BYN7 U5EHL8 A0A151XF28 A0A0L0C6S2 A0A023EVP6 A0A2P8XXV8 A0A336MYI8 A0A336L6S7 A0A1L8DDY7 A0A336MYE0 A0A067RKF7 A0A336N4K8 A0A1S4FDF4 Q176H5 Q7QBV8 A0A1Q3FHN8 A0A1Q3FH17 A0A182HTD9 A0A182LFI7 A0A182P863 B0WZD9 W8B4W6 A0A182HF16 A0A0A1XJ05 A0A0A1XCP3 N6TDV8 A0A182U8Y5 U4UHE5 A0A182MYK8 A0A182UME3 Q9VC29 B4PV34 Q29BI6 A0A182JU99 B4QTT8 A0A182FU53 A0A0K8UCR2 A0A336L4K1 W5J1Z2 A0A3B0JIL0 A0A182QFM7 A0A1B0DRA4 B3LV12 A0A182XEJ3 Q9U6Q2 A0A182UAG7 Q16ED9 A0A084WHU1 A0A182UN25 Q7PV84 A0A182HQ40 A0A182XNB9 A0A1B6IYY2 A0A182R5D4
PDB
5DA0
E-value=7.53824e-12,
Score=172
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
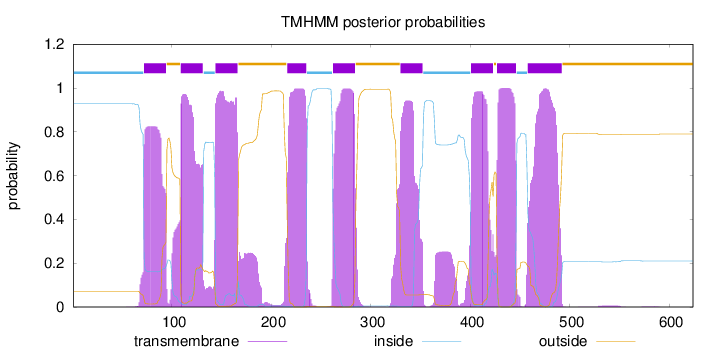
Length:
624
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
205.84235
Exp number, first 60 AAs:
0.00223
Total prob of N-in:
0.92834
inside
1 - 71
TMhelix
72 - 94
outside
95 - 108
TMhelix
109 - 131
inside
132 - 143
TMhelix
144 - 166
outside
167 - 215
TMhelix
216 - 235
inside
236 - 261
TMhelix
262 - 284
outside
285 - 329
TMhelix
330 - 352
inside
353 - 400
TMhelix
401 - 423
outside
424 - 426
TMhelix
427 - 446
inside
447 - 457
TMhelix
458 - 492
outside
493 - 624
Population Genetic Test Statistics
Pi
168.995611
Theta
160.949884
Tajima's D
-0.823304
CLR
0.295681
CSRT
0.171841407929604
Interpretation
Uncertain
