Pre Gene Modal
BGIBMGA003931
Annotation
PREDICTED:_WD_repeat_domain_phosphoinositide-interacting_protein_2_[Papilio_machaon]
Location in the cell
Extracellular Reliability : 1.102 PlasmaMembrane Reliability : 1.385
Sequence
CDS
ATGAACACACTGTCCGTTTCCATCGCTCCAATGAGTCTGGGAGGAGTTACAAACACTGAGGGATCGAATGCAGGCGGAATATTCGTTCAGTTTAATCAAGATTGCACGTCATTGGTGGCGGGCAGCAGCACCGGATATCACTTATTTGCTCTAACTCCTGATGATGGTATCGAAGAAATATATGCGAGTCGCAGCGGACAGGAGACTTGTCTCGTCGACCGTCTCTTCAGCAGCTCGCTGGTGGCTGTTGTCACAGTATCCGCTCCCAGGAAGCTGATAGTGTGCCACTACAAGAAGGGCACCGAGATCTGCAACTACATCTACAGCAACACCATCCTCGCCGTCAAACTGAACCGCTCGAGGTTGATCGTGTGCCTCGAGGAGTCGCTGCACATCCACAACATCCGGGACATGAAGATCCTGCACACGATCCGCGACACGCCGCCCAACCCGCGCGGCCTGTGCGCGCTGTCGCCCTGCGTGGAGCACTGCTACGTGGCGTACCCCGGCTCCAGCGCGGTGGGCGAGGTGCAGATCTTCGACGCGGTGCACCTCAACGCCAAGTGCGCCATCAGCGCGCACGAGAGCCCCGTGGCGGCGCTGGCCTGGTCCTCCTGCGGGCGGCGCCTGGCCACCGCCTCGGAGCGCGGCACCGTCATCCGCGTGTTCGCCGTGCCCGACCGCACGCGCCTGTTCGAGTTCCGACGCGGCGTCAAGCGCTGCGTCACCATCGCCTGCCTCGCCTTCAGCAGCTGCGGCCTGTACCTCGCCGCCACCTCCAACACGGAGACCGTGCACGTGTTCAAGCTGCAGGAGGGCGGCGCGGGCGAGGAGGGCGGCGCGGGCGAGACGGCGGGCGCCGCGGCGGCGGGCGAGGCGGACACGTGGCTGGGCTGGCTGTCGGCCGCGGTGTCGGCGGGCGCGGGCTACCTGCCGGCGCACGTGGCCGACGTGCTGGCGCAGGGCCGCGCCTTCGCCGCCGCGCGCCTGCCCGCCACCGGCCACCGCGCCGTCGCCGCCGTCACGAGAGTGTCGCGCAGTCCGCGGCTGCTGGTGGCCACGTCGGCCGGGGTGCTGTACGTGTACGCGCTGGACGCGGCGGAGGGCGGCGAGTGCGCGCTGCTGCGGCAGCACCGCTTCCTGGCGCCGGAGGAGGCGCCGGGCTCCGCGGACCCGCAAGTGGCGACGGGCAACAGTTACGCGGGCGCGCTGCGCGGCCGGGACCCCGCCGCCATGACAGCTGCAGGTGTGTGTGCGTCAGATTCAGAGCGCGGCCGGGAGCTGGGCGCCGCGCTCGAGCCCTCCCCGCCGCGGGGACCCTTCGACCTCCGCGACCCCGACCACTTCCCCCCCATCGCCGTCAGGGCACCGCAGCACTAG
Protein
MNTLSVSIAPMSLGGVTNTEGSNAGGIFVQFNQDCTSLVAGSSTGYHLFALTPDDGIEEIYASRSGQETCLVDRLFSSSLVAVVTVSAPRKLIVCHYKKGTEICNYIYSNTILAVKLNRSRLIVCLEESLHIHNIRDMKILHTIRDTPPNPRGLCALSPCVEHCYVAYPGSSAVGEVQIFDAVHLNAKCAISAHESPVAALAWSSCGRRLATASERGTVIRVFAVPDRTRLFEFRRGVKRCVTIACLAFSSCGLYLAATSNTETVHVFKLQEGGAGEEGGAGETAGAAAAGEADTWLGWLSAAVSAGAGYLPAHVADVLAQGRAFAAARLPATGHRAVAAVTRVSRSPRLLVATSAGVLYVYALDAAEGGECALLRQHRFLAPEEAPGSADPQVATGNSYAGALRGRDPAAMTAAGVCASDSERGRELGAALEPSPPRGPFDLRDPDHFPPIAVRAPQH
Summary
Uniprot
A0A2A4JN63
A0A2H1V4U1
A0A212F942
A0A194QFD9
A0A194R3J1
A0A1Y1JZZ7
+ More
A0A0P6D6L6 A0A0P6D3J7 A0A0P6EQM4 A0A0P6DT66 A0A0C9R9R8 A0A2J7PKT3 A0A2A3EC32 A0A088AER0 A0A0P5AJ58 A0A0P5YQM7 A0A0P5U3S2 A0A195CDC6 A0A0P6D3K0 A0A0P5CRK9 A0A0N8CM12 A0A158P376 A0A0P5HE49 A0A1B6C5R2 A0A0P5HC23 A0A195BIQ1 A0A195DN47 A0A195EZE4 A0A2J7PKU0 A0A1B6DMR4 A0A0P6H867 A0A0N8EIM9 A0A0P5U2Y3 A0A151WZK0 A0A0P5S6E2 A0A0P5LRJ1 A0A0P4YUV4 E2AP41 V5I979 A0A067RCU6 A0A0P5K4U8 A0A0P6GH03 A0A0P6G999 A0A0C9QC46 A0A2J7PKT1 A0A0P5GQX7 A0A0P6E9E2 A0A0P5NY14 A0A0P6ADV0 A0A0P5UHK8 A0A0P5JG11 A0A0P5SCS4 A0A0P6E4U6 A0A0P5KPX6 A0A0P5RK93 A0A0P5RKC8 A0A0P5BNB5 A0A0P5JFX5 A0A0P5S323 A0A0P5PRP6 A0A0A9YHB1 A0A0P4VZU5 A0A1Y1K144 K7IY06 D6WVG2 A0A0A9W107 A0A3B3D2T7 A0A2J7PKT9 A0A3B4V421 A0A154P646 A0A3B3TYY6 A0A3B4YCE4 A0A3P8V8P6 A0A3B5MJB7 A0A232F1L2 H2SCP5 A0A1A8FIG9 A0A1S3JDL4 A0A3Q3AX91 A0A3Q1HP25 A0A3P9KI98 H2MEQ1 A0A3B3YJE5 M3ZFR3 A0A087YBQ5 A0A3P9PZJ6 A0A3Q3L448 A0A2I4BKS7 A0A1A8D9A4 A0A3B3H438 A0A210QTQ5 A0A1A8KV28 A0A1A8B5W1 A0A3P9IFL8 A0A3Q3D849 A0A2U9CS19 A0A2U9CPV7 A0A1A8QWP0
A0A0P6D6L6 A0A0P6D3J7 A0A0P6EQM4 A0A0P6DT66 A0A0C9R9R8 A0A2J7PKT3 A0A2A3EC32 A0A088AER0 A0A0P5AJ58 A0A0P5YQM7 A0A0P5U3S2 A0A195CDC6 A0A0P6D3K0 A0A0P5CRK9 A0A0N8CM12 A0A158P376 A0A0P5HE49 A0A1B6C5R2 A0A0P5HC23 A0A195BIQ1 A0A195DN47 A0A195EZE4 A0A2J7PKU0 A0A1B6DMR4 A0A0P6H867 A0A0N8EIM9 A0A0P5U2Y3 A0A151WZK0 A0A0P5S6E2 A0A0P5LRJ1 A0A0P4YUV4 E2AP41 V5I979 A0A067RCU6 A0A0P5K4U8 A0A0P6GH03 A0A0P6G999 A0A0C9QC46 A0A2J7PKT1 A0A0P5GQX7 A0A0P6E9E2 A0A0P5NY14 A0A0P6ADV0 A0A0P5UHK8 A0A0P5JG11 A0A0P5SCS4 A0A0P6E4U6 A0A0P5KPX6 A0A0P5RK93 A0A0P5RKC8 A0A0P5BNB5 A0A0P5JFX5 A0A0P5S323 A0A0P5PRP6 A0A0A9YHB1 A0A0P4VZU5 A0A1Y1K144 K7IY06 D6WVG2 A0A0A9W107 A0A3B3D2T7 A0A2J7PKT9 A0A3B4V421 A0A154P646 A0A3B3TYY6 A0A3B4YCE4 A0A3P8V8P6 A0A3B5MJB7 A0A232F1L2 H2SCP5 A0A1A8FIG9 A0A1S3JDL4 A0A3Q3AX91 A0A3Q1HP25 A0A3P9KI98 H2MEQ1 A0A3B3YJE5 M3ZFR3 A0A087YBQ5 A0A3P9PZJ6 A0A3Q3L448 A0A2I4BKS7 A0A1A8D9A4 A0A3B3H438 A0A210QTQ5 A0A1A8KV28 A0A1A8B5W1 A0A3P9IFL8 A0A3Q3D849 A0A2U9CS19 A0A2U9CPV7 A0A1A8QWP0
Pubmed
EMBL
NWSH01000930
PCG73505.1
ODYU01000676
SOQ35853.1
AGBW02009651
OWR50248.1
+ More
KQ459144 KPJ03660.1 KQ460779 KPJ12373.1 GEZM01099224 GEZM01099223 JAV53430.1 GDIQ01083305 JAN11432.1 GDIP01044014 GDIQ01083307 LRGB01000389 JAM59701.1 JAN11430.1 KZS19446.1 GDIQ01059216 JAN35521.1 GDIQ01073161 JAN21576.1 GBYB01004960 JAG74727.1 NEVH01024531 PNF16942.1 KZ288301 PBC28852.1 GDIP01198244 JAJ25158.1 GDIP01055065 JAM48650.1 GDIP01118550 JAL85164.1 KQ977935 KYM98710.1 GDIP01044013 GDIQ01083306 JAM59702.1 JAN11431.1 GDIP01166728 JAJ56674.1 GDIP01118551 JAL85163.1 ADTU01007771 ADTU01007772 ADTU01007773 ADTU01007774 GDIQ01228900 JAK22825.1 GEDC01028462 JAS08836.1 GDIQ01228901 JAK22824.1 KQ976467 KYM84188.1 KQ980713 KYN14251.1 KQ981905 KYN33588.1 PNF16948.1 GEDC01010349 JAS26949.1 GDIQ01035482 JAN59255.1 GDIQ01023036 JAN71701.1 GDIP01118549 JAL85165.1 KQ982649 KYQ53105.1 GDIQ01098153 JAL53573.1 GDIQ01166281 JAK85444.1 GDIP01226930 JAI96471.1 GL441452 EFN64777.1 GALX01003405 JAB65061.1 KK852784 KDR16597.1 GDIQ01189266 JAK62459.1 GDIQ01045768 JAN48969.1 GDIQ01038273 JAN56464.1 GBYB01000824 JAG70591.1 PNF16944.1 GDIQ01237663 JAK14062.1 GDIQ01067470 JAN27267.1 GDIQ01136183 JAL15543.1 GDIP01031009 JAM72706.1 GDIP01114156 GDIQ01067469 JAL89558.1 JAN27268.1 GDIQ01200522 JAK51203.1 GDIP01141557 JAL62157.1 GDIQ01073160 JAN21577.1 GDIQ01186420 JAK65305.1 GDIQ01118645 JAL33081.1 GDIQ01099505 JAL52221.1 GDIP01187195 JAJ36207.1 GDIQ01200568 JAK51157.1 GDIQ01099506 JAL52220.1 GDIQ01130839 JAL20887.1 GBHO01042076 GBHO01014679 GBRD01015383 GDHC01010303 JAG01528.1 JAG28925.1 JAG50443.1 JAQ08326.1 GDKW01001542 JAI55053.1 GEZM01099228 GEZM01099225 JAV53425.1 KQ971357 EFA08562.1 GBHO01042080 GBHO01042070 GBRD01015382 JAG01524.1 JAG01534.1 JAG50444.1 PNF16946.1 KQ434823 KZC07322.1 NNAY01001254 OXU24604.1 HAEB01012034 HAEC01002519 SBQ58561.1 AYCK01015927 HADZ01019029 HAEA01002375 SBQ30855.1 NEDP02001973 OWF52090.1 HAED01010050 HAEE01016340 SBR36390.1 HADY01024412 HAEJ01000619 SBP62897.1 CP026261 AWP18619.1 AWP18618.1 HAEH01013693 HAEI01017117 SBR98056.1
KQ459144 KPJ03660.1 KQ460779 KPJ12373.1 GEZM01099224 GEZM01099223 JAV53430.1 GDIQ01083305 JAN11432.1 GDIP01044014 GDIQ01083307 LRGB01000389 JAM59701.1 JAN11430.1 KZS19446.1 GDIQ01059216 JAN35521.1 GDIQ01073161 JAN21576.1 GBYB01004960 JAG74727.1 NEVH01024531 PNF16942.1 KZ288301 PBC28852.1 GDIP01198244 JAJ25158.1 GDIP01055065 JAM48650.1 GDIP01118550 JAL85164.1 KQ977935 KYM98710.1 GDIP01044013 GDIQ01083306 JAM59702.1 JAN11431.1 GDIP01166728 JAJ56674.1 GDIP01118551 JAL85163.1 ADTU01007771 ADTU01007772 ADTU01007773 ADTU01007774 GDIQ01228900 JAK22825.1 GEDC01028462 JAS08836.1 GDIQ01228901 JAK22824.1 KQ976467 KYM84188.1 KQ980713 KYN14251.1 KQ981905 KYN33588.1 PNF16948.1 GEDC01010349 JAS26949.1 GDIQ01035482 JAN59255.1 GDIQ01023036 JAN71701.1 GDIP01118549 JAL85165.1 KQ982649 KYQ53105.1 GDIQ01098153 JAL53573.1 GDIQ01166281 JAK85444.1 GDIP01226930 JAI96471.1 GL441452 EFN64777.1 GALX01003405 JAB65061.1 KK852784 KDR16597.1 GDIQ01189266 JAK62459.1 GDIQ01045768 JAN48969.1 GDIQ01038273 JAN56464.1 GBYB01000824 JAG70591.1 PNF16944.1 GDIQ01237663 JAK14062.1 GDIQ01067470 JAN27267.1 GDIQ01136183 JAL15543.1 GDIP01031009 JAM72706.1 GDIP01114156 GDIQ01067469 JAL89558.1 JAN27268.1 GDIQ01200522 JAK51203.1 GDIP01141557 JAL62157.1 GDIQ01073160 JAN21577.1 GDIQ01186420 JAK65305.1 GDIQ01118645 JAL33081.1 GDIQ01099505 JAL52221.1 GDIP01187195 JAJ36207.1 GDIQ01200568 JAK51157.1 GDIQ01099506 JAL52220.1 GDIQ01130839 JAL20887.1 GBHO01042076 GBHO01014679 GBRD01015383 GDHC01010303 JAG01528.1 JAG28925.1 JAG50443.1 JAQ08326.1 GDKW01001542 JAI55053.1 GEZM01099228 GEZM01099225 JAV53425.1 KQ971357 EFA08562.1 GBHO01042080 GBHO01042070 GBRD01015382 JAG01524.1 JAG01534.1 JAG50444.1 PNF16946.1 KQ434823 KZC07322.1 NNAY01001254 OXU24604.1 HAEB01012034 HAEC01002519 SBQ58561.1 AYCK01015927 HADZ01019029 HAEA01002375 SBQ30855.1 NEDP02001973 OWF52090.1 HAED01010050 HAEE01016340 SBR36390.1 HADY01024412 HAEJ01000619 SBP62897.1 CP026261 AWP18619.1 AWP18618.1 HAEH01013693 HAEI01017117 SBR98056.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000076858
UP000235965
+ More
UP000242457 UP000005203 UP000078542 UP000005205 UP000078540 UP000078492 UP000078541 UP000075809 UP000000311 UP000027135 UP000002358 UP000007266 UP000261560 UP000261420 UP000076502 UP000261500 UP000261360 UP000265120 UP000261380 UP000215335 UP000005226 UP000085678 UP000264800 UP000265040 UP000265180 UP000001038 UP000261480 UP000002852 UP000028760 UP000242638 UP000261640 UP000192220 UP000242188 UP000265200 UP000264820 UP000246464
UP000242457 UP000005203 UP000078542 UP000005205 UP000078540 UP000078492 UP000078541 UP000075809 UP000000311 UP000027135 UP000002358 UP000007266 UP000261560 UP000261420 UP000076502 UP000261500 UP000261360 UP000265120 UP000261380 UP000215335 UP000005226 UP000085678 UP000264800 UP000265040 UP000265180 UP000001038 UP000261480 UP000002852 UP000028760 UP000242638 UP000261640 UP000192220 UP000242188 UP000265200 UP000264820 UP000246464
Pfam
PF17906 HTH_48
Interpro
SUPFAM
SSF50978
SSF50978
Gene 3D
ProteinModelPortal
A0A2A4JN63
A0A2H1V4U1
A0A212F942
A0A194QFD9
A0A194R3J1
A0A1Y1JZZ7
+ More
A0A0P6D6L6 A0A0P6D3J7 A0A0P6EQM4 A0A0P6DT66 A0A0C9R9R8 A0A2J7PKT3 A0A2A3EC32 A0A088AER0 A0A0P5AJ58 A0A0P5YQM7 A0A0P5U3S2 A0A195CDC6 A0A0P6D3K0 A0A0P5CRK9 A0A0N8CM12 A0A158P376 A0A0P5HE49 A0A1B6C5R2 A0A0P5HC23 A0A195BIQ1 A0A195DN47 A0A195EZE4 A0A2J7PKU0 A0A1B6DMR4 A0A0P6H867 A0A0N8EIM9 A0A0P5U2Y3 A0A151WZK0 A0A0P5S6E2 A0A0P5LRJ1 A0A0P4YUV4 E2AP41 V5I979 A0A067RCU6 A0A0P5K4U8 A0A0P6GH03 A0A0P6G999 A0A0C9QC46 A0A2J7PKT1 A0A0P5GQX7 A0A0P6E9E2 A0A0P5NY14 A0A0P6ADV0 A0A0P5UHK8 A0A0P5JG11 A0A0P5SCS4 A0A0P6E4U6 A0A0P5KPX6 A0A0P5RK93 A0A0P5RKC8 A0A0P5BNB5 A0A0P5JFX5 A0A0P5S323 A0A0P5PRP6 A0A0A9YHB1 A0A0P4VZU5 A0A1Y1K144 K7IY06 D6WVG2 A0A0A9W107 A0A3B3D2T7 A0A2J7PKT9 A0A3B4V421 A0A154P646 A0A3B3TYY6 A0A3B4YCE4 A0A3P8V8P6 A0A3B5MJB7 A0A232F1L2 H2SCP5 A0A1A8FIG9 A0A1S3JDL4 A0A3Q3AX91 A0A3Q1HP25 A0A3P9KI98 H2MEQ1 A0A3B3YJE5 M3ZFR3 A0A087YBQ5 A0A3P9PZJ6 A0A3Q3L448 A0A2I4BKS7 A0A1A8D9A4 A0A3B3H438 A0A210QTQ5 A0A1A8KV28 A0A1A8B5W1 A0A3P9IFL8 A0A3Q3D849 A0A2U9CS19 A0A2U9CPV7 A0A1A8QWP0
A0A0P6D6L6 A0A0P6D3J7 A0A0P6EQM4 A0A0P6DT66 A0A0C9R9R8 A0A2J7PKT3 A0A2A3EC32 A0A088AER0 A0A0P5AJ58 A0A0P5YQM7 A0A0P5U3S2 A0A195CDC6 A0A0P6D3K0 A0A0P5CRK9 A0A0N8CM12 A0A158P376 A0A0P5HE49 A0A1B6C5R2 A0A0P5HC23 A0A195BIQ1 A0A195DN47 A0A195EZE4 A0A2J7PKU0 A0A1B6DMR4 A0A0P6H867 A0A0N8EIM9 A0A0P5U2Y3 A0A151WZK0 A0A0P5S6E2 A0A0P5LRJ1 A0A0P4YUV4 E2AP41 V5I979 A0A067RCU6 A0A0P5K4U8 A0A0P6GH03 A0A0P6G999 A0A0C9QC46 A0A2J7PKT1 A0A0P5GQX7 A0A0P6E9E2 A0A0P5NY14 A0A0P6ADV0 A0A0P5UHK8 A0A0P5JG11 A0A0P5SCS4 A0A0P6E4U6 A0A0P5KPX6 A0A0P5RK93 A0A0P5RKC8 A0A0P5BNB5 A0A0P5JFX5 A0A0P5S323 A0A0P5PRP6 A0A0A9YHB1 A0A0P4VZU5 A0A1Y1K144 K7IY06 D6WVG2 A0A0A9W107 A0A3B3D2T7 A0A2J7PKT9 A0A3B4V421 A0A154P646 A0A3B3TYY6 A0A3B4YCE4 A0A3P8V8P6 A0A3B5MJB7 A0A232F1L2 H2SCP5 A0A1A8FIG9 A0A1S3JDL4 A0A3Q3AX91 A0A3Q1HP25 A0A3P9KI98 H2MEQ1 A0A3B3YJE5 M3ZFR3 A0A087YBQ5 A0A3P9PZJ6 A0A3Q3L448 A0A2I4BKS7 A0A1A8D9A4 A0A3B3H438 A0A210QTQ5 A0A1A8KV28 A0A1A8B5W1 A0A3P9IFL8 A0A3Q3D849 A0A2U9CS19 A0A2U9CPV7 A0A1A8QWP0
PDB
5LTG
E-value=4.15262e-42,
Score=432
Ontologies
GO
PANTHER
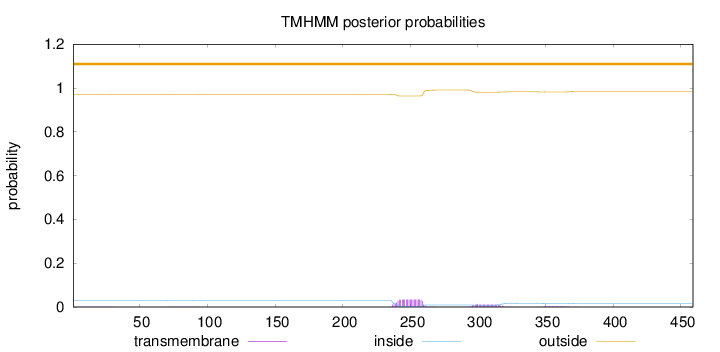
Topology
Length:
459
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.04018
Exp number, first 60 AAs:
0.01306
Total prob of N-in:
0.02979
outside
1 - 459
Population Genetic Test Statistics
Pi
138.739814
Theta
152.717378
Tajima's D
-1.07737
CLR
376.954315
CSRT
0.123993800309984
Interpretation
Uncertain
