Gene
KWMTBOMO10633
Pre Gene Modal
BGIBMGA003937
Annotation
PREDICTED:_prominin-like_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 1.713
Sequence
CDS
ATGAATAAAATCAGCCAGCAACTGATCCTTGATATGTCGGACGTTATGGACCTGGCGACTCCTAATTACTCGACACCCATAATTAACACAACTTATGTGGCCAGTCTGCAATTCGACATGCGAGCCATGGGCCCGCTATACAACTCCACACATATGATCATCGACGCTATCGCCAATAAACAAGCTTATCCTGAAGGAATAGTGTCCGTGTCCGACGGTCACTTGGACATCGCGTCACCGCGAGACAATTGGAAGGTGCTGTTAGCGCATTACGCCGGGCCCACCGCCATCGTAGTAGTCACGGCACTGGTGATTGCTGCTCTGCCACTAGCCGGGTTATTCTGGTGCTGCTGCTACTGGTGCCGCGTGGGACGACGCCGGAGACCCTTCGACCGCAAATATGACGCCTGTCTCAAAGGCATCCTTGCCATACTCCTCATTGGATTGCTCACATTATTCTTGTTCGGAGTGGTCTGCGCGTTCGCGACGGACTCTCAAGTGGAGGACGGGATGGCGGACGCTCCGGACTCGCTGCGGGACGGCATCCGGGACGCCCAGGAGTTCCTCAACGCGACGCAGGCGCACGCCCGCTGGCTGCTCGTCGTCAACTACAGGGAGCTCAAGGACGCTCTGAACGCGGTGCTCCGCACGAGCGGCATGACTGTGGCGCTGAAGCTGGGCGAGTTCTCGCACGCGGCGTCGGTGAGCGCGCTCAGCCGCATGGTGGCGCGGCTGGACGACGTGCGCGCCGACCTGCGCGCCGTGCACGCGCTCACCGACGCGCTGCGACACCACGCCGACGCGCTCAACGCAGGTCTCCGCAAGGTGAAGAACCAGCTGCTCCAGACCCTGGCGCGCTGCGACCAGCCCAACTGCAAGGCTCTGCAGAACAAGTACAAGCTGGGCCAGCTGGACACGGAGATACACTACAGTCAGATGCCGGACGTGTCTCCGCTACTCAGCAGCGTCTCCGTGCTCCTCGAGGGACACATCCAGGAGGAGGTGGCCGCCGGTCAGCAAGTGTTCAGGGACATACAGTACGGGATCCGGCGCTCCATCGACACCCACATACCCGACGTGCAGAACGCCATCGAGGAGACCGGTCGCAAGCTGGAGGGCATCGCGGAGCAGATCACGCAGCTGGCCGGCAACGCGAGCGCGCGGCTGGCGAGCGACGGCGGCGGCGCGGCGGACGCGCTGCGGAGCCTGCACGCGCGCTACGGGCCCTACCGCCGCCACGCCGGGCTGGCGGCCGCGCTCGCGCTCCTAGCGATAACGTTGCTGCTGGTGTGGGGGCTGGTGTGCGGCGTGTGCGGCAAGCGCCCCGACGCGTACGGCGCCGCGGACTGCTGCAACAAGGGCGCCGGCGCGCGCTGCCTGCTCTGGTACCGCGCTCTCTCTGAACTGTCCACTTTGCGAAACCAAAGCGGCGTGGGCGCGACGCTGGCGGCGGGCGGCGCGGTGGCGCTGGTGCTGCTGGTGCACTTCGCCGCGGGGCTCGCCGCGCAGCGCTTCGTGTGCGACCCGCTCACAGAGCCCCGCGGTAACCGTCTGTTCGAGGACCTGGAGAAGTTCGTCGACCTGGAGAAGATGCTGTTCAACGAGCGGCTAGACCCCAACTTCAACATGACGTCCGTGCTGCTGCAGTGCCACATGAACCACACCATATACGAGACGCTGCAGCTGCGGCGCGTGTACGACGTGGAGGAGCTGCGGCGCGCGCTGGGGGCCGCCGTGGGGGCGCGCGTGCGGTCCCTGCGCGCCCGCTACCCGCAGGAGGCGGCGCGCGGCCGCGTCACGCTGCTGCGGGACCCCGCCAAGCTCAAGCTGACCGAGCTGGCCAACACGCAGCTCTCGGACTTCGACTTCGACAAGATCCTGGATGCCCTGGAGAGCAACCTGACGACGCTGTCGCTGGAGGAGCTGGGCGAGCGGCTGACGAGCGCGGCGGGCGCGCGGGGGCTGGCGGGGTCGCGGGAGGGGGCGGACCTGCGCCGCGCCGCCGCCGCGCTGGCGCAGCTGCACGCCGACCTGCTGCTGCCCATGCGCAACTACACGCTCCAGCTCAACCGGACGGCGACAAAACTCCGCGACGAGCTGCGGTTCAACCAGTCGTCGCTGAAGGCGGCCATCTTGTACCACCTCCACGAGACCAACGAGGCCGAGGACTTCCTCAACAACCAGGGGCCGCAGCTGCTGGAGAACATCACGGCGGCGTTCGCGGCGGTGGTGGAGCGCGAGGTGGGCGCGTACGGCGAGCGCCTGTCCGCCGCCGCCGCGCGCCACGTGGGCCGCTGCGGTCCGCTCTCCGCCGGATTCAACGCCACGCGCGACGCGCTCTGCACCAAGCTGCTGCTGCCCGCGGTGAGTGCCGCCCCCGCCCCGCCCCGCCCGCACCCCGCCCCCGACCCCCCGCCTCACCGCGCCTGTGCCCCGCAGAACGGCTACTGGATGTCGCTGGCGTGGGGCCTGCTGGTGCTGGTGCCGGTGCTGGTGGTGGCGCAGCGGCTGGCCGGGCTGTACCTGCAGGCCGACCCCTACCCCGGACCGCTAGTCGAGGCCGAATATTTGTATGACGCGTACGCGGACCGCGATAACGTACCCCTCGCCAACAGTGGGTCAAAACGCAGTACGCTGGACATCGAGGGTAAGATTGCGGAGTGGAGGGACCGGGCCGTCCCCTCCGCCCCCCCGCCCCACTCGCCTCCCCCGTCGCCCCCGCACTCTCGCACGCAGCTCCTGCTCGCGGGGGAGGAGGTCGTGTGCCAGCTGGTGGCCCCGCCCCCCGGACTCATCGACGACCTCAAGACTCGACTCGACGCTAAAGACGAGGACAACGAGTCCGGAATACACGAGTCCGAGTAA
Protein
MNKISQQLILDMSDVMDLATPNYSTPIINTTYVASLQFDMRAMGPLYNSTHMIIDAIANKQAYPEGIVSVSDGHLDIASPRDNWKVLLAHYAGPTAIVVVTALVIAALPLAGLFWCCCYWCRVGRRRRPFDRKYDACLKGILAILLIGLLTLFLFGVVCAFATDSQVEDGMADAPDSLRDGIRDAQEFLNATQAHARWLLVVNYRELKDALNAVLRTSGMTVALKLGEFSHAASVSALSRMVARLDDVRADLRAVHALTDALRHHADALNAGLRKVKNQLLQTLARCDQPNCKALQNKYKLGQLDTEIHYSQMPDVSPLLSSVSVLLEGHIQEEVAAGQQVFRDIQYGIRRSIDTHIPDVQNAIEETGRKLEGIAEQITQLAGNASARLASDGGGAADALRSLHARYGPYRRHAGLAAALALLAITLLLVWGLVCGVCGKRPDAYGAADCCNKGAGARCLLWYRALSELSTLRNQSGVGATLAAGGAVALVLLVHFAAGLAAQRFVCDPLTEPRGNRLFEDLEKFVDLEKMLFNERLDPNFNMTSVLLQCHMNHTIYETLQLRRVYDVEELRRALGAAVGARVRSLRARYPQEAARGRVTLLRDPAKLKLTELANTQLSDFDFDKILDALESNLTTLSLEELGERLTSAAGARGLAGSREGADLRRAAAALAQLHADLLLPMRNYTLQLNRTATKLRDELRFNQSSLKAAILYHLHETNEAEDFLNNQGPQLLENITAAFAAVVEREVGAYGERLSAAAARHVGRCGPLSAGFNATRDALCTKLLLPAVSAAPAPPRPHPAPDPPPHRACAPQNGYWMSLAWGLLVLVPVLVVAQRLAGLYLQADPYPGPLVEAEYLYDAYADRDNVPLANSGSKRSTLDIEGKIAEWRDRAVPSAPPPHSPPPSPPHSRTQLLLAGEEVVCQLVAPPPGLIDDLKTRLDAKDEDNESGIHESE
Summary
Uniprot
A0A2W1BPV0
A0A2A4JA74
A0A194QDP4
A0A212F924
A0A2H1WKS7
A0A0C9RXU2
+ More
A0A0C9R600 A0A088AR64 A0A026W1W3 A0A0M8ZWH5 K7IYK1 A0A151WI20 A0A0C9R554 A0A0L7REI1 A0A2A3EA35 A0A232F9P1 A0A3L8DMJ7 A0A158NJI2 A0A151HZH9 F4WDR5 A0A154PPD0 A0A1B6EEW6 A0A1B6DYQ5 A0A0J7P3N1 E2AWX0 A0A2S2QLV8 A0A151IN77 A0A224XJ65 A0A2H8TQS2 U5ETW0 A0A1B6HH08 J9JRA3 A0A2S2QEB5 E2BAM1 A0A336M658 A0A336M8P2 A0A1I8N4I7 A0A0L0CCD5 A0A182JC95 A0A1I8PIR5 A0A182NIR2 A0A2J7PK32 A0A2M4A3N0 A0A182Q3K6 A0A2M4CNM4 Q7Q6R9 A0A182H8V0 A0A182V1X5 A0A182PMI8 A0A182FCB6 A0A182HWN4 A0A182GU57 A0A182XMS7 A0A182K8E4 A0A0P5N4V6 W5JI92 Q16LQ3 A0A1I8N4H8 A0A1I8N4I1 A0A1I8N4I9 A0A0P5DB46 E9FV88 A0A0P5W2V2 A0A0P5YZ42 A0A0P6AHW3 A0A0P5ZK93 A0A0N8CAM9 A0A0P6HYC3 A0A1J1J8M8 T1P8V7 A0A0P6AEC0 A0A2M4A3H8 A0A0T6AU22 A0A2J7PK29 A0A0P5E7P4 A0A0P5SC99 A0A0P5S9U8 A0A1Y1NFW8 A0A0N8AZQ3 A0A0P4ZGU5 A0A0N8A353 A0A0P5AR46 A0A0P5ZGP8 A0A0P5Y2H6 A0A0P5TAG3 A0A0P5E5F7 A0A0P5VLN5 A0A0P6HGH4 A0A3R7PCC5 A0A0N8CD04
A0A0C9R600 A0A088AR64 A0A026W1W3 A0A0M8ZWH5 K7IYK1 A0A151WI20 A0A0C9R554 A0A0L7REI1 A0A2A3EA35 A0A232F9P1 A0A3L8DMJ7 A0A158NJI2 A0A151HZH9 F4WDR5 A0A154PPD0 A0A1B6EEW6 A0A1B6DYQ5 A0A0J7P3N1 E2AWX0 A0A2S2QLV8 A0A151IN77 A0A224XJ65 A0A2H8TQS2 U5ETW0 A0A1B6HH08 J9JRA3 A0A2S2QEB5 E2BAM1 A0A336M658 A0A336M8P2 A0A1I8N4I7 A0A0L0CCD5 A0A182JC95 A0A1I8PIR5 A0A182NIR2 A0A2J7PK32 A0A2M4A3N0 A0A182Q3K6 A0A2M4CNM4 Q7Q6R9 A0A182H8V0 A0A182V1X5 A0A182PMI8 A0A182FCB6 A0A182HWN4 A0A182GU57 A0A182XMS7 A0A182K8E4 A0A0P5N4V6 W5JI92 Q16LQ3 A0A1I8N4H8 A0A1I8N4I1 A0A1I8N4I9 A0A0P5DB46 E9FV88 A0A0P5W2V2 A0A0P5YZ42 A0A0P6AHW3 A0A0P5ZK93 A0A0N8CAM9 A0A0P6HYC3 A0A1J1J8M8 T1P8V7 A0A0P6AEC0 A0A2M4A3H8 A0A0T6AU22 A0A2J7PK29 A0A0P5E7P4 A0A0P5SC99 A0A0P5S9U8 A0A1Y1NFW8 A0A0N8AZQ3 A0A0P4ZGU5 A0A0N8A353 A0A0P5AR46 A0A0P5ZGP8 A0A0P5Y2H6 A0A0P5TAG3 A0A0P5E5F7 A0A0P5VLN5 A0A0P6HGH4 A0A3R7PCC5 A0A0N8CD04
Pubmed
EMBL
KZ149989
PZC75634.1
NWSH01002377
PCG68434.1
KQ459144
KPJ03673.1
+ More
AGBW02009651 OWR50242.1 ODYU01009333 SOQ53683.1 GBYB01014055 JAG83822.1 GBYB01003400 JAG73167.1 KK107503 EZA49566.1 KQ435837 KOX71476.1 KQ983097 KYQ47499.1 GBYB01003090 JAG72857.1 KQ414608 KOC69352.1 KZ288313 PBC28344.1 NNAY01000655 OXU27158.1 QOIP01000006 RLU21443.1 ADTU01017793 ADTU01017794 ADTU01017795 ADTU01017796 ADTU01017797 ADTU01017798 ADTU01017799 ADTU01017800 ADTU01017801 ADTU01017802 KQ976688 KYM78129.1 GL888090 EGI67678.1 KQ435010 KZC13741.1 GEDC01000821 JAS36477.1 GEDC01006513 JAS30785.1 LBMM01000187 KMR04544.1 GL443425 EFN62090.1 GGMS01009534 MBY78737.1 KQ976958 KYN06796.1 GFTR01008355 JAW08071.1 GFXV01004739 MBW16544.1 GANO01002606 JAB57265.1 GECU01033724 GECU01018006 JAS73982.1 JAS89700.1 ABLF02035220 ABLF02035221 GGMS01006657 MBY75860.1 GL446793 EFN87247.1 UFQT01000614 SSX25762.1 UFQT01000253 SSX22408.1 JRES01000611 KNC29872.1 NEVH01024942 PNF16690.1 GGFK01002009 MBW35330.1 AXCN02000419 GGFL01002715 MBW66893.1 AAAB01008960 EAA10743.3 JXUM01029560 KQ560856 KXJ80616.1 APCN01001511 JXUM01018352 JXUM01018353 JXUM01018354 JXUM01018355 KQ560510 KXJ82058.1 GDIQ01147264 JAL04462.1 ADMH02001414 ETN62615.1 CH477897 EAT35263.1 GDIP01163914 JAJ59488.1 GL732525 EFX89151.1 GDIP01105874 JAL97840.1 GDIP01051238 JAM52477.1 GDIP01029546 JAM74169.1 GDIP01043841 JAM59874.1 GDIQ01098427 JAL53299.1 GDIQ01028049 JAN66688.1 CVRI01000075 CRL08743.1 KA645122 AFP59751.1 GDIP01043840 JAM59875.1 GGFK01002026 MBW35347.1 LJIG01022816 KRT78569.1 PNF16691.1 GDIP01150468 JAJ72934.1 GDIP01141528 JAL62186.1 GDIP01142577 JAL61137.1 GEZM01006962 GEZM01006961 JAV95525.1 GDIQ01227035 JAK24690.1 GDIP01213019 JAJ10383.1 GDIP01187113 JAJ36289.1 GDIP01195165 JAJ28237.1 GDIP01043842 JAM59873.1 GDIP01064893 JAM38822.1 GDIP01130330 JAL73384.1 GDIP01147819 JAJ75583.1 GDIP01112978 JAL90736.1 GDIQ01028803 JAN65934.1 QCYY01000925 ROT81731.1 GDIQ01091827 JAL59899.1
AGBW02009651 OWR50242.1 ODYU01009333 SOQ53683.1 GBYB01014055 JAG83822.1 GBYB01003400 JAG73167.1 KK107503 EZA49566.1 KQ435837 KOX71476.1 KQ983097 KYQ47499.1 GBYB01003090 JAG72857.1 KQ414608 KOC69352.1 KZ288313 PBC28344.1 NNAY01000655 OXU27158.1 QOIP01000006 RLU21443.1 ADTU01017793 ADTU01017794 ADTU01017795 ADTU01017796 ADTU01017797 ADTU01017798 ADTU01017799 ADTU01017800 ADTU01017801 ADTU01017802 KQ976688 KYM78129.1 GL888090 EGI67678.1 KQ435010 KZC13741.1 GEDC01000821 JAS36477.1 GEDC01006513 JAS30785.1 LBMM01000187 KMR04544.1 GL443425 EFN62090.1 GGMS01009534 MBY78737.1 KQ976958 KYN06796.1 GFTR01008355 JAW08071.1 GFXV01004739 MBW16544.1 GANO01002606 JAB57265.1 GECU01033724 GECU01018006 JAS73982.1 JAS89700.1 ABLF02035220 ABLF02035221 GGMS01006657 MBY75860.1 GL446793 EFN87247.1 UFQT01000614 SSX25762.1 UFQT01000253 SSX22408.1 JRES01000611 KNC29872.1 NEVH01024942 PNF16690.1 GGFK01002009 MBW35330.1 AXCN02000419 GGFL01002715 MBW66893.1 AAAB01008960 EAA10743.3 JXUM01029560 KQ560856 KXJ80616.1 APCN01001511 JXUM01018352 JXUM01018353 JXUM01018354 JXUM01018355 KQ560510 KXJ82058.1 GDIQ01147264 JAL04462.1 ADMH02001414 ETN62615.1 CH477897 EAT35263.1 GDIP01163914 JAJ59488.1 GL732525 EFX89151.1 GDIP01105874 JAL97840.1 GDIP01051238 JAM52477.1 GDIP01029546 JAM74169.1 GDIP01043841 JAM59874.1 GDIQ01098427 JAL53299.1 GDIQ01028049 JAN66688.1 CVRI01000075 CRL08743.1 KA645122 AFP59751.1 GDIP01043840 JAM59875.1 GGFK01002026 MBW35347.1 LJIG01022816 KRT78569.1 PNF16691.1 GDIP01150468 JAJ72934.1 GDIP01141528 JAL62186.1 GDIP01142577 JAL61137.1 GEZM01006962 GEZM01006961 JAV95525.1 GDIQ01227035 JAK24690.1 GDIP01213019 JAJ10383.1 GDIP01187113 JAJ36289.1 GDIP01195165 JAJ28237.1 GDIP01043842 JAM59873.1 GDIP01064893 JAM38822.1 GDIP01130330 JAL73384.1 GDIP01147819 JAJ75583.1 GDIP01112978 JAL90736.1 GDIQ01028803 JAN65934.1 QCYY01000925 ROT81731.1 GDIQ01091827 JAL59899.1
Proteomes
UP000218220
UP000053268
UP000007151
UP000005203
UP000053097
UP000053105
+ More
UP000002358 UP000075809 UP000053825 UP000242457 UP000215335 UP000279307 UP000005205 UP000078540 UP000007755 UP000076502 UP000036403 UP000000311 UP000078542 UP000007819 UP000008237 UP000095301 UP000037069 UP000075880 UP000095300 UP000075884 UP000235965 UP000075886 UP000007062 UP000069940 UP000249989 UP000075903 UP000075885 UP000069272 UP000075840 UP000076407 UP000075881 UP000000673 UP000008820 UP000000305 UP000183832 UP000283509
UP000002358 UP000075809 UP000053825 UP000242457 UP000215335 UP000279307 UP000005205 UP000078540 UP000007755 UP000076502 UP000036403 UP000000311 UP000078542 UP000007819 UP000008237 UP000095301 UP000037069 UP000075880 UP000095300 UP000075884 UP000235965 UP000075886 UP000007062 UP000069940 UP000249989 UP000075903 UP000075885 UP000069272 UP000075840 UP000076407 UP000075881 UP000000673 UP000008820 UP000000305 UP000183832 UP000283509
Pfam
PF05478 Prominin
Interpro
IPR008795
Prominin
ProteinModelPortal
A0A2W1BPV0
A0A2A4JA74
A0A194QDP4
A0A212F924
A0A2H1WKS7
A0A0C9RXU2
+ More
A0A0C9R600 A0A088AR64 A0A026W1W3 A0A0M8ZWH5 K7IYK1 A0A151WI20 A0A0C9R554 A0A0L7REI1 A0A2A3EA35 A0A232F9P1 A0A3L8DMJ7 A0A158NJI2 A0A151HZH9 F4WDR5 A0A154PPD0 A0A1B6EEW6 A0A1B6DYQ5 A0A0J7P3N1 E2AWX0 A0A2S2QLV8 A0A151IN77 A0A224XJ65 A0A2H8TQS2 U5ETW0 A0A1B6HH08 J9JRA3 A0A2S2QEB5 E2BAM1 A0A336M658 A0A336M8P2 A0A1I8N4I7 A0A0L0CCD5 A0A182JC95 A0A1I8PIR5 A0A182NIR2 A0A2J7PK32 A0A2M4A3N0 A0A182Q3K6 A0A2M4CNM4 Q7Q6R9 A0A182H8V0 A0A182V1X5 A0A182PMI8 A0A182FCB6 A0A182HWN4 A0A182GU57 A0A182XMS7 A0A182K8E4 A0A0P5N4V6 W5JI92 Q16LQ3 A0A1I8N4H8 A0A1I8N4I1 A0A1I8N4I9 A0A0P5DB46 E9FV88 A0A0P5W2V2 A0A0P5YZ42 A0A0P6AHW3 A0A0P5ZK93 A0A0N8CAM9 A0A0P6HYC3 A0A1J1J8M8 T1P8V7 A0A0P6AEC0 A0A2M4A3H8 A0A0T6AU22 A0A2J7PK29 A0A0P5E7P4 A0A0P5SC99 A0A0P5S9U8 A0A1Y1NFW8 A0A0N8AZQ3 A0A0P4ZGU5 A0A0N8A353 A0A0P5AR46 A0A0P5ZGP8 A0A0P5Y2H6 A0A0P5TAG3 A0A0P5E5F7 A0A0P5VLN5 A0A0P6HGH4 A0A3R7PCC5 A0A0N8CD04
A0A0C9R600 A0A088AR64 A0A026W1W3 A0A0M8ZWH5 K7IYK1 A0A151WI20 A0A0C9R554 A0A0L7REI1 A0A2A3EA35 A0A232F9P1 A0A3L8DMJ7 A0A158NJI2 A0A151HZH9 F4WDR5 A0A154PPD0 A0A1B6EEW6 A0A1B6DYQ5 A0A0J7P3N1 E2AWX0 A0A2S2QLV8 A0A151IN77 A0A224XJ65 A0A2H8TQS2 U5ETW0 A0A1B6HH08 J9JRA3 A0A2S2QEB5 E2BAM1 A0A336M658 A0A336M8P2 A0A1I8N4I7 A0A0L0CCD5 A0A182JC95 A0A1I8PIR5 A0A182NIR2 A0A2J7PK32 A0A2M4A3N0 A0A182Q3K6 A0A2M4CNM4 Q7Q6R9 A0A182H8V0 A0A182V1X5 A0A182PMI8 A0A182FCB6 A0A182HWN4 A0A182GU57 A0A182XMS7 A0A182K8E4 A0A0P5N4V6 W5JI92 Q16LQ3 A0A1I8N4H8 A0A1I8N4I1 A0A1I8N4I9 A0A0P5DB46 E9FV88 A0A0P5W2V2 A0A0P5YZ42 A0A0P6AHW3 A0A0P5ZK93 A0A0N8CAM9 A0A0P6HYC3 A0A1J1J8M8 T1P8V7 A0A0P6AEC0 A0A2M4A3H8 A0A0T6AU22 A0A2J7PK29 A0A0P5E7P4 A0A0P5SC99 A0A0P5S9U8 A0A1Y1NFW8 A0A0N8AZQ3 A0A0P4ZGU5 A0A0N8A353 A0A0P5AR46 A0A0P5ZGP8 A0A0P5Y2H6 A0A0P5TAG3 A0A0P5E5F7 A0A0P5VLN5 A0A0P6HGH4 A0A3R7PCC5 A0A0N8CD04
Ontologies
GO
PANTHER
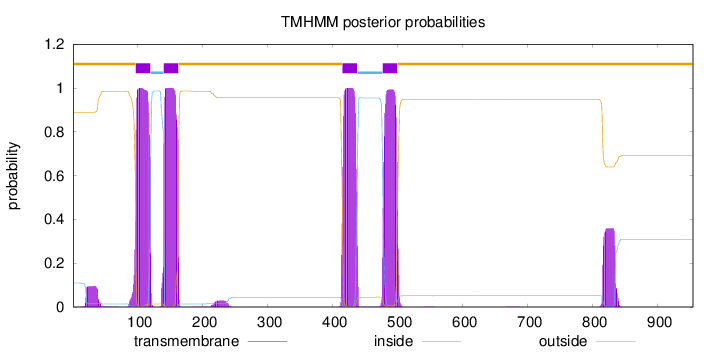
Topology
Length:
954
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
100.793940000001
Exp number, first 60 AAs:
1.90333
Total prob of N-in:
0.11018
outside
1 - 96
TMhelix
97 - 119
inside
120 - 139
TMhelix
140 - 162
outside
163 - 414
TMhelix
415 - 437
inside
438 - 476
TMhelix
477 - 499
outside
500 - 954
Population Genetic Test Statistics
Pi
195.340816
Theta
173.64678
Tajima's D
0.273796
CLR
0.552888
CSRT
0.451377431128444
Interpretation
Uncertain
