Gene
KWMTBOMO10631
Pre Gene Modal
BGIBMGA003960
Annotation
PREDICTED:_sodium-independent_sulfate_anion_transporter-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.513
Sequence
CDS
ATGACGAAATACAAACTGAACAACACTATTCGTTCTTCGCCTGAAGTCACACTGACGGCTTACATTGCTGACGACGCTAAGAAACCTGACGAGAAGAAGTACGAGATCGCAGTATTGGACAGCAATCAAAACGACAGTGCCGTGAACGAAAACAACCGCAGTCTCGGGTCGAAGATATGTCACGTTTGTCCTCACCCGTGTGAGTTCGCGAAACACTCCGACCGACTGGCCGCCTCGCTGGTGAAGCAAGAGTGCGACAAATTCGGCTTGAACGAGCTCAGAGTTGAGGGCTTAGAGAAGAACGTGTGTCTGTGCGAGAGATACAACGGAGGCTGCAAACTATGCCCTCGTCCGCTGAATGCCAATTTGAATCTTACGCTGAACGGGCGAGAGTCGTGCTTGGCATGTCACAAGGCGACCGATTCCTACTCTAATACAAATGGATCATATCCAGGAAGTCTGTTCACGAAGACCCCCGTTAGCTGGAGGAGGAAGGCAGCTAGACGCGCCAGAGCGTGTTGCTCCAAGAAAATGCTGCTCCGAAGAGTTCCTATACTGCAGTGGTTACCCAACTACACTTGGCGCAGCGGACTGTCTGATACTATTGCGGGTATAACGGTGGGTCTCACAGTCATACCTCAGGCTATAGCTTATGCCGGAGTGGCCGGCTTGCCGCCGCAGTATGGACTCTACTCCTCATTTATGGCCTGCTTCGTTTACACGATCTTCGGTTCAGTAAAGGACTCGGCTATAGGTCCCACCGCTATCGCCGCTATACTGACGCGGGAAAATCTCCACGGCCTGGGACCAGAATTTGCTGTTCTACTGGCCTTTCTGTCTGGGTGCGTGGAGCTGGTCATGGGAATGTTGCAATTAGGTTTTCTGATTGACTTCATCAGCGGTCCGGTCTCCGTTGGGTTCACATCTGCCGCTGCCATCATCATAGCCACTACTCAGGTTAAAGACATCCTCGGGTTAAGCTTCCCTGGTGGCAAGTTTATTCAGGTGTGGACGGGGATATACGAACACATCGGTGAGACTCGACTCTGGGACACTGTGCTCGGTGTAACTTGTATCGTTGTACTGCTTCTGTTGAGGAAAGTCAAGGATATAAGCGTGAAGGTCGATGAGAGTTCAGGCAAGACCAGGCGTCTGCGTAAGGCCGCCTCGCAGACACTGTGGCTCGTGTCCACCACGCGGAACATCCTGGTGGTGCTGGCCTGTGCATCATTGGCCTACTACTTCGACACGAGGCAGTTGCAACCTTTCGTTTTAACTGGTGAAGTAAAGGCGGGGCTACCACAGATCTCCCCCCCACCATTCTCCGCCACGGTCGGTAACCACACGTACACGTTTGTGGAGATGGCATCCACCCTCGGATCAGCGATATTCGTGGTGCCACTCCTCTCTATTTTAGAGAATATAGCGTTGGCTAAAGTTTTTTCGGAGGGTAAGTACGTGGACGCGACGCAAGAGATGCTGGCTCTGGGCGTGTGCAACATGGTGTCGGCGCTGGTGGACAGTATGCCGGTGTCGGGGGCGCTGTCCCGCGGCGCCGTCAACCACGCCTCCGGGGTCGCCACCACCGCCTCGGGCCTCTGGGCCGGCGCGCTGGTGCTGCTCGCGCTACAGTATTTCACGCCGTATTTCTACTACATTCCGAAAGCGTCATTGGCTGCCGTCATCATTGCTGCTGTGGTCTTCATGATGGAACTGCACGTCTTCAAACCTATATGGAGGACGAAAAAAATCGATATAATCCCCGCGGTGGTGACGTTCGGCTCTTGTCTCATGCTGCGACTGGAGCTTGGCATCGTCATAGGCATCGGCGTTAACTTGCTGTTCCTGCTGTATGCCTCCGCGAGACCCTCCGTCAAGGTCACGCGGGCCACCTCTGCGAGCGGCGTGGAGCACGTGGTGGCGGCGGTGGGCAGCGCGCTGTCGTTCCCGGCGGCGGAGTTCGTGCGGCGCGCGCTGCACAAGGCGGCGCCGCGCCTGCCGCTCGTGGTGGACGCGACGCACGTCAGGGCGGCAGACTTCACCGCCGCAGAGGGTATCAAGGCGTTAATTGAAGATTTCGAGTCTCGTGGGCAAACGCTGATCTTCTACAACGTGTCTCCGTCGCTGGGCAGCGTGCTGCAGGGCGTGCGGCCCCGGAGACTGCTGGTGTGCTCCTCCGCCACTCAGCTGGACCTGCTGCTCGTGCAAGCCACGCAGCAAAGCCTCACCAACTCATAG
Protein
MTKYKLNNTIRSSPEVTLTAYIADDAKKPDEKKYEIAVLDSNQNDSAVNENNRSLGSKICHVCPHPCEFAKHSDRLAASLVKQECDKFGLNELRVEGLEKNVCLCERYNGGCKLCPRPLNANLNLTLNGRESCLACHKATDSYSNTNGSYPGSLFTKTPVSWRRKAARRARACCSKKMLLRRVPILQWLPNYTWRSGLSDTIAGITVGLTVIPQAIAYAGVAGLPPQYGLYSSFMACFVYTIFGSVKDSAIGPTAIAAILTRENLHGLGPEFAVLLAFLSGCVELVMGMLQLGFLIDFISGPVSVGFTSAAAIIIATTQVKDILGLSFPGGKFIQVWTGIYEHIGETRLWDTVLGVTCIVVLLLLRKVKDISVKVDESSGKTRRLRKAASQTLWLVSTTRNILVVLACASLAYYFDTRQLQPFVLTGEVKAGLPQISPPPFSATVGNHTYTFVEMASTLGSAIFVVPLLSILENIALAKVFSEGKYVDATQEMLALGVCNMVSALVDSMPVSGALSRGAVNHASGVATTASGLWAGALVLLALQYFTPYFYYIPKASLAAVIIAAVVFMMELHVFKPIWRTKKIDIIPAVVTFGSCLMLRLELGIVIGIGVNLLFLLYASARPSVKVTRATSASGVEHVVAAVGSALSFPAAEFVRRALHKAAPRLPLVVDATHVRAADFTAAEGIKALIEDFESRGQTLIFYNVSPSLGSVLQGVRPRRLLVCSSATQLDLLLVQATQQSLTNS
Summary
Similarity
Belongs to the SLC26A/SulP transporter (TC 2.A.53) family.
Uniprot
A0A2A4IZ56
A0A2W1BKM4
A0A212F938
H9J373
A0A194QFF3
A0A194R3K6
+ More
A0A2H1WIT1 A0A067RKA6 A0A1W4X674 D2A570 E0VH04 A0A195BDI2 A0A3L8DCX7 A0A0J7NVE5 A0A026WK90 F4WGT4 A0A1B6BYH8 A0A151JV11 E2C4G5 A0A0T6BGS6 A0A069DV45 A0A224X9U9 A0A023F3Y6 A0A0P4VQI8 T1IES7 A0A088A5L7 A0A2A3E4T2 A0A151JSC7 E1ZW33 A0A232EP20 K7IZ34 A0A0L7R2P2 A0A2S2R092 J9JU03 A0A2H8TFV1 A0A0A9Z6Y1 A0A0A9Z2J6 A0A146M619 A0A146KSN0 A0A151WP73 A0A0M8ZQ79 U4UYB3 N6U798 A0A310SR24 A0A2J7QAE0 A0A2J7QAB9 D6WUN9 A0A067RPK8 A0A1B6CML3 E0VCB0 A0A1W4XLX0 A0A067RKF7 A0A2P8XXV8 V5H1U5 A0A310SI74 A0A182QWG3 E2BP27 A0A1B6DIL8 A0A0C9Q9Z3 A0A0K8TQT4 A0A154PR24 A0A194PIX2 A0A1L8DEP9 A0A182IXP3 A0A026X0F7 A0A182NGD0 A0A182W2Q5 A0A1B0DRA4 A0A2A3EBU0 A0A088AR89 E2ANG6 A0A1B0CLE2 A0A195FKC5 A0A182M4V4 N6TDV8 K7IYL9 A0A069DWI5 A0A182F6K0 A0A2M3ZFE6 A0A2M4A5J3 A0A1B0DR61 W5JUZ8 U4UHE5 A0A195C7D8 A0A2M4BGI5 A0A2M4BGE8 A0A1W4XEM9 A0A182RY98 A0A1W4XEN4 A0A158NTY7 A0A151I4S0 A0A2H1V6V8 A0A1B0CTN7 A0A195EMB3 V9IDC5 Q17AX2
A0A2H1WIT1 A0A067RKA6 A0A1W4X674 D2A570 E0VH04 A0A195BDI2 A0A3L8DCX7 A0A0J7NVE5 A0A026WK90 F4WGT4 A0A1B6BYH8 A0A151JV11 E2C4G5 A0A0T6BGS6 A0A069DV45 A0A224X9U9 A0A023F3Y6 A0A0P4VQI8 T1IES7 A0A088A5L7 A0A2A3E4T2 A0A151JSC7 E1ZW33 A0A232EP20 K7IZ34 A0A0L7R2P2 A0A2S2R092 J9JU03 A0A2H8TFV1 A0A0A9Z6Y1 A0A0A9Z2J6 A0A146M619 A0A146KSN0 A0A151WP73 A0A0M8ZQ79 U4UYB3 N6U798 A0A310SR24 A0A2J7QAE0 A0A2J7QAB9 D6WUN9 A0A067RPK8 A0A1B6CML3 E0VCB0 A0A1W4XLX0 A0A067RKF7 A0A2P8XXV8 V5H1U5 A0A310SI74 A0A182QWG3 E2BP27 A0A1B6DIL8 A0A0C9Q9Z3 A0A0K8TQT4 A0A154PR24 A0A194PIX2 A0A1L8DEP9 A0A182IXP3 A0A026X0F7 A0A182NGD0 A0A182W2Q5 A0A1B0DRA4 A0A2A3EBU0 A0A088AR89 E2ANG6 A0A1B0CLE2 A0A195FKC5 A0A182M4V4 N6TDV8 K7IYL9 A0A069DWI5 A0A182F6K0 A0A2M3ZFE6 A0A2M4A5J3 A0A1B0DR61 W5JUZ8 U4UHE5 A0A195C7D8 A0A2M4BGI5 A0A2M4BGE8 A0A1W4XEM9 A0A182RY98 A0A1W4XEN4 A0A158NTY7 A0A151I4S0 A0A2H1V6V8 A0A1B0CTN7 A0A195EMB3 V9IDC5 Q17AX2
Pubmed
EMBL
NWSH01004565
PCG64919.1
KZ149989
PZC75632.1
AGBW02009651
OWR50240.1
+ More
BABH01036699 BABH01036700 BABH01036701 BABH01036702 KQ459144 KPJ03675.1 KQ460779 KPJ12388.1 ODYU01008954 SOQ52983.1 KK852421 KDR24301.1 KQ971361 EFA05122.1 DS235156 EEB12660.1 KQ976514 KYM82269.1 QOIP01000010 RLU18002.1 LBMM01001414 KMQ96365.1 KK107167 EZA56467.1 GL888147 EGI66553.1 GEDC01030992 JAS06306.1 KQ981724 KYN37236.1 GL452471 EFN77149.1 LJIG01000412 KRT86528.1 GBGD01000966 JAC87923.1 GFTR01007289 JAW09137.1 GBBI01002547 JAC16165.1 GDKW01003168 JAI53427.1 ACPB03015825 KZ288400 PBC26292.1 KQ978538 KYN30257.1 GL434761 EFN74608.1 NNAY01003020 OXU20103.1 KQ414665 KOC65145.1 GGMS01014215 MBY83418.1 ABLF02032283 ABLF02032286 ABLF02032290 ABLF02032292 GFXV01001159 MBW12964.1 GBHO01006039 JAG37565.1 GBHO01006041 GBHO01006040 GBRD01002176 JAG37563.1 JAG37564.1 JAG63645.1 GDHC01003426 JAQ15203.1 GDHC01019650 JAP98978.1 KQ982877 KYQ49702.1 KQ435912 KOX68890.1 KB632424 ERL95346.1 APGK01040018 APGK01040019 KB740975 ENN76516.1 KQ760116 OAD61939.1 NEVH01016331 PNF25529.1 PNF25526.1 KQ971363 EFA07819.1 KK852498 KDR22565.1 GEDC01022690 JAS14608.1 DS235051 EEB10996.1 KDR24302.1 PYGN01001181 PSN36843.1 GALX01001643 JAB66823.1 KQ759878 OAD62248.1 AXCN02001614 GL449511 EFN82619.1 GEDC01011801 JAS25497.1 GBYB01011218 GBYB01011219 JAG80985.1 JAG80986.1 GDAI01000881 JAI16722.1 KQ435010 KZC13728.1 KQ459603 KPI92998.1 GFDF01009163 JAV04921.1 KK107046 QOIP01000004 EZA61752.1 RLU23357.1 AJVK01019779 KZ288289 PBC29243.1 GL441216 EFN65015.1 AJWK01017158 AJWK01017159 AJWK01017160 AJWK01017161 AJWK01017162 AJWK01017163 KQ981522 KYN40712.1 AXCM01000024 APGK01041487 APGK01041488 APGK01041489 APGK01041490 KB740994 ENN75938.1 GBGD01000827 JAC88062.1 GGFM01006501 MBW27252.1 GGFK01002679 MBW36000.1 AJVK01019712 ADMH02000060 ETN67956.1 KB632356 ERL93394.1 KQ978143 KYM96789.1 GGFJ01002996 MBW52137.1 GGFJ01002995 MBW52136.1 ADTU01026210 KQ976445 KYM86093.1 ODYU01000799 SOQ36122.1 AJWK01027790 KQ978691 KYN29266.1 JR039342 AEY58647.1 CH477329 EAT43399.1
BABH01036699 BABH01036700 BABH01036701 BABH01036702 KQ459144 KPJ03675.1 KQ460779 KPJ12388.1 ODYU01008954 SOQ52983.1 KK852421 KDR24301.1 KQ971361 EFA05122.1 DS235156 EEB12660.1 KQ976514 KYM82269.1 QOIP01000010 RLU18002.1 LBMM01001414 KMQ96365.1 KK107167 EZA56467.1 GL888147 EGI66553.1 GEDC01030992 JAS06306.1 KQ981724 KYN37236.1 GL452471 EFN77149.1 LJIG01000412 KRT86528.1 GBGD01000966 JAC87923.1 GFTR01007289 JAW09137.1 GBBI01002547 JAC16165.1 GDKW01003168 JAI53427.1 ACPB03015825 KZ288400 PBC26292.1 KQ978538 KYN30257.1 GL434761 EFN74608.1 NNAY01003020 OXU20103.1 KQ414665 KOC65145.1 GGMS01014215 MBY83418.1 ABLF02032283 ABLF02032286 ABLF02032290 ABLF02032292 GFXV01001159 MBW12964.1 GBHO01006039 JAG37565.1 GBHO01006041 GBHO01006040 GBRD01002176 JAG37563.1 JAG37564.1 JAG63645.1 GDHC01003426 JAQ15203.1 GDHC01019650 JAP98978.1 KQ982877 KYQ49702.1 KQ435912 KOX68890.1 KB632424 ERL95346.1 APGK01040018 APGK01040019 KB740975 ENN76516.1 KQ760116 OAD61939.1 NEVH01016331 PNF25529.1 PNF25526.1 KQ971363 EFA07819.1 KK852498 KDR22565.1 GEDC01022690 JAS14608.1 DS235051 EEB10996.1 KDR24302.1 PYGN01001181 PSN36843.1 GALX01001643 JAB66823.1 KQ759878 OAD62248.1 AXCN02001614 GL449511 EFN82619.1 GEDC01011801 JAS25497.1 GBYB01011218 GBYB01011219 JAG80985.1 JAG80986.1 GDAI01000881 JAI16722.1 KQ435010 KZC13728.1 KQ459603 KPI92998.1 GFDF01009163 JAV04921.1 KK107046 QOIP01000004 EZA61752.1 RLU23357.1 AJVK01019779 KZ288289 PBC29243.1 GL441216 EFN65015.1 AJWK01017158 AJWK01017159 AJWK01017160 AJWK01017161 AJWK01017162 AJWK01017163 KQ981522 KYN40712.1 AXCM01000024 APGK01041487 APGK01041488 APGK01041489 APGK01041490 KB740994 ENN75938.1 GBGD01000827 JAC88062.1 GGFM01006501 MBW27252.1 GGFK01002679 MBW36000.1 AJVK01019712 ADMH02000060 ETN67956.1 KB632356 ERL93394.1 KQ978143 KYM96789.1 GGFJ01002996 MBW52137.1 GGFJ01002995 MBW52136.1 ADTU01026210 KQ976445 KYM86093.1 ODYU01000799 SOQ36122.1 AJWK01027790 KQ978691 KYN29266.1 JR039342 AEY58647.1 CH477329 EAT43399.1
Proteomes
UP000218220
UP000007151
UP000005204
UP000053268
UP000053240
UP000027135
+ More
UP000192223 UP000007266 UP000009046 UP000078540 UP000279307 UP000036403 UP000053097 UP000007755 UP000078541 UP000008237 UP000015103 UP000005203 UP000242457 UP000078492 UP000000311 UP000215335 UP000002358 UP000053825 UP000007819 UP000075809 UP000053105 UP000030742 UP000019118 UP000235965 UP000245037 UP000075886 UP000076502 UP000075880 UP000075884 UP000075920 UP000092462 UP000092461 UP000075883 UP000069272 UP000000673 UP000078542 UP000075900 UP000005205 UP000008820
UP000192223 UP000007266 UP000009046 UP000078540 UP000279307 UP000036403 UP000053097 UP000007755 UP000078541 UP000008237 UP000015103 UP000005203 UP000242457 UP000078492 UP000000311 UP000215335 UP000002358 UP000053825 UP000007819 UP000075809 UP000053105 UP000030742 UP000019118 UP000235965 UP000245037 UP000075886 UP000076502 UP000075880 UP000075884 UP000075920 UP000092462 UP000092461 UP000075883 UP000069272 UP000000673 UP000078542 UP000075900 UP000005205 UP000008820
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4IZ56
A0A2W1BKM4
A0A212F938
H9J373
A0A194QFF3
A0A194R3K6
+ More
A0A2H1WIT1 A0A067RKA6 A0A1W4X674 D2A570 E0VH04 A0A195BDI2 A0A3L8DCX7 A0A0J7NVE5 A0A026WK90 F4WGT4 A0A1B6BYH8 A0A151JV11 E2C4G5 A0A0T6BGS6 A0A069DV45 A0A224X9U9 A0A023F3Y6 A0A0P4VQI8 T1IES7 A0A088A5L7 A0A2A3E4T2 A0A151JSC7 E1ZW33 A0A232EP20 K7IZ34 A0A0L7R2P2 A0A2S2R092 J9JU03 A0A2H8TFV1 A0A0A9Z6Y1 A0A0A9Z2J6 A0A146M619 A0A146KSN0 A0A151WP73 A0A0M8ZQ79 U4UYB3 N6U798 A0A310SR24 A0A2J7QAE0 A0A2J7QAB9 D6WUN9 A0A067RPK8 A0A1B6CML3 E0VCB0 A0A1W4XLX0 A0A067RKF7 A0A2P8XXV8 V5H1U5 A0A310SI74 A0A182QWG3 E2BP27 A0A1B6DIL8 A0A0C9Q9Z3 A0A0K8TQT4 A0A154PR24 A0A194PIX2 A0A1L8DEP9 A0A182IXP3 A0A026X0F7 A0A182NGD0 A0A182W2Q5 A0A1B0DRA4 A0A2A3EBU0 A0A088AR89 E2ANG6 A0A1B0CLE2 A0A195FKC5 A0A182M4V4 N6TDV8 K7IYL9 A0A069DWI5 A0A182F6K0 A0A2M3ZFE6 A0A2M4A5J3 A0A1B0DR61 W5JUZ8 U4UHE5 A0A195C7D8 A0A2M4BGI5 A0A2M4BGE8 A0A1W4XEM9 A0A182RY98 A0A1W4XEN4 A0A158NTY7 A0A151I4S0 A0A2H1V6V8 A0A1B0CTN7 A0A195EMB3 V9IDC5 Q17AX2
A0A2H1WIT1 A0A067RKA6 A0A1W4X674 D2A570 E0VH04 A0A195BDI2 A0A3L8DCX7 A0A0J7NVE5 A0A026WK90 F4WGT4 A0A1B6BYH8 A0A151JV11 E2C4G5 A0A0T6BGS6 A0A069DV45 A0A224X9U9 A0A023F3Y6 A0A0P4VQI8 T1IES7 A0A088A5L7 A0A2A3E4T2 A0A151JSC7 E1ZW33 A0A232EP20 K7IZ34 A0A0L7R2P2 A0A2S2R092 J9JU03 A0A2H8TFV1 A0A0A9Z6Y1 A0A0A9Z2J6 A0A146M619 A0A146KSN0 A0A151WP73 A0A0M8ZQ79 U4UYB3 N6U798 A0A310SR24 A0A2J7QAE0 A0A2J7QAB9 D6WUN9 A0A067RPK8 A0A1B6CML3 E0VCB0 A0A1W4XLX0 A0A067RKF7 A0A2P8XXV8 V5H1U5 A0A310SI74 A0A182QWG3 E2BP27 A0A1B6DIL8 A0A0C9Q9Z3 A0A0K8TQT4 A0A154PR24 A0A194PIX2 A0A1L8DEP9 A0A182IXP3 A0A026X0F7 A0A182NGD0 A0A182W2Q5 A0A1B0DRA4 A0A2A3EBU0 A0A088AR89 E2ANG6 A0A1B0CLE2 A0A195FKC5 A0A182M4V4 N6TDV8 K7IYL9 A0A069DWI5 A0A182F6K0 A0A2M3ZFE6 A0A2M4A5J3 A0A1B0DR61 W5JUZ8 U4UHE5 A0A195C7D8 A0A2M4BGI5 A0A2M4BGE8 A0A1W4XEM9 A0A182RY98 A0A1W4XEN4 A0A158NTY7 A0A151I4S0 A0A2H1V6V8 A0A1B0CTN7 A0A195EMB3 V9IDC5 Q17AX2
PDB
5DA0
E-value=3.78546e-10,
Score=158
Ontologies
GO
PANTHER
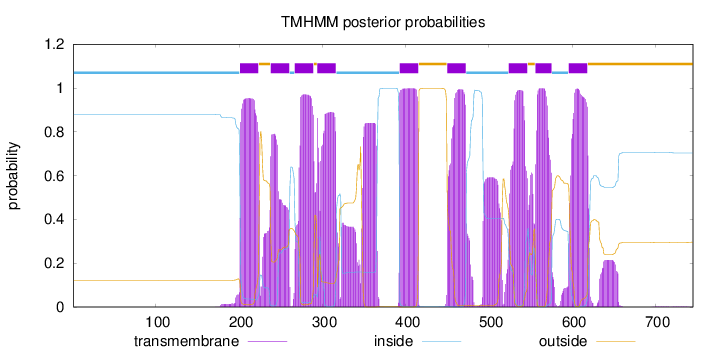
Topology
Subcellular location
Membrane
Length:
745
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
230.60851
Exp number, first 60 AAs:
0
Total prob of N-in:
0.87962
inside
1 - 200
TMhelix
201 - 223
outside
224 - 237
TMhelix
238 - 260
inside
261 - 266
TMhelix
267 - 289
outside
290 - 293
TMhelix
294 - 316
inside
317 - 392
TMhelix
393 - 415
outside
416 - 449
TMhelix
450 - 472
inside
473 - 523
TMhelix
524 - 546
outside
547 - 555
TMhelix
556 - 575
inside
576 - 595
TMhelix
596 - 618
outside
619 - 745
Population Genetic Test Statistics
Pi
160.208835
Theta
164.617464
Tajima's D
-0.855415
CLR
1.501934
CSRT
0.171391430428479
Interpretation
Uncertain
