Gene
KWMTBOMO10626 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA003957
Annotation
cytochrome_P450_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.403
Sequence
CDS
ATGATCACTTTAATCTGGTTGGGTGTACTTCTGGTGACACTGACGTTACACCTCAGGAAGGTGTACTCAAGGTTCAAGGATTATGGCGTGAACCACTTCACGCCGATCCCGGTGTTAGGAAACGCTGGTCCAATAACAGTGCGACTTCGGCACGTGGCTGAGGACTTCGATATGGTGTATAAAGCTTTTCCAGAGGACAGGTTCACAGGCAGATTTGACTTGCTGCGGCCCACGGTTATCATCAAAGATTTAGACCTCATAAAGCAGATCACTATAAAAGACTTTGAGCATTTCCTGGACCACAGAGCTTTGGTTGATGACACCGCTGACCCGTTCTTTGGCAGAAACCTATTTTCTTTGAGAGGTCAAGAATGGAAAGACATGCGGTCCACGTTGAGTCCTGCTTTCACCAGCTCCAAGATGCGTGGAATGGTCCCTTTTATGGTGGAAGTTAATAACCAGATGATTGATATGATTAAAAAAAAAATTGTGGCTAATGCGGATGGCTACCTCGACTGTGAAGGTAAAGACCTGACCACTCGCTACGCTAATGACGTCATAGCATCCTGTGCATTCGGCGTCAAGGTGGACTCCCATACCAATGAGGAAAATCAGTTCTATCTTATGGGTAGGGACATGGCGGATTTTGGATTCCGAAAGATTATGGTATTTCTCGGTTACTCTTCGTTCCCGAAATTAATGAAGAAATTCAACGCCAAACTTCTGTCTGATGAGACTGGACATTTCTTCACCGATTTAGTTCTCAGAACGATGGAAGACCGTGAGGTTAAAGAAATTGTGAGACCTGATATGATTCATTTACTGATGGAGGCAAAACAAGGCAAACTTTCTTATGATGAAAAAAGTACTAAAGAGGCTGACACTGGATTTGCAACAGTCGAAGAATCCGATGTCGGAAAAAAGACTATCAATAGAATCTGGTCCAACACCGATTTGATTGCCCAAGCAACCCTCTTCTTCGTGGCTGGCTTTGAGACTATTTCGTCGGCGATGTCGTTTGCACTTCACGAGTTGGCTCTAAACCCTGAAATTCAGGACCGTCTGGTGCAAGAGATCAAGGAAAACTACGCGAAGACTGGGGGGAAATTCGATTTTAACTGTATTCAAGATTTGACTTACATGGATATGTTTGTGTCGGAGGTACTTAGATTGTGGACTCCGGTAGTCGGCATGGATCGTCTCTGCGTCAAAGATTACAATCTGGGAAGAGCGAACAAAAACGCCACTAAAGACTTTATCCTCCGCAAAGGCGAAGGATTGTCTATCCCGACCTGGTCAATCCATCACAATCCTGAATACTACCCCGAACCCTATAAATTTGACCCAGAGCGCTTCTCCGAGGAAAACAAACGGAACATCAAACCGTTCACTTATCTACCATTCGGGACGGGTCCGAGGAATTGTATCGGTTCAAGGTTTGCTCTTTGCGAGGTGAAAGTCATGTTATACCAGCTCCTGCAGCAGATCGAAGTTCTTCCGAGCGACAAGACTAAGGTTCGAGCGAAATTGGCGAAGGATACCTTCAACGTCAAGATCGAAGGAGGCCACTGGATCAGACTCAAGCTTAGGGATTAG
Protein
MITLIWLGVLLVTLTLHLRKVYSRFKDYGVNHFTPIPVLGNAGPITVRLRHVAEDFDMVYKAFPEDRFTGRFDLLRPTVIIKDLDLIKQITIKDFEHFLDHRALVDDTADPFFGRNLFSLRGQEWKDMRSTLSPAFTSSKMRGMVPFMVEVNNQMIDMIKKKIVANADGYLDCEGKDLTTRYANDVIASCAFGVKVDSHTNEENQFYLMGRDMADFGFRKIMVFLGYSSFPKLMKKFNAKLLSDETGHFFTDLVLRTMEDREVKEIVRPDMIHLLMEAKQGKLSYDEKSTKEADTGFATVEESDVGKKTINRIWSNTDLIAQATLFFVAGFETISSAMSFALHELALNPEIQDRLVQEIKENYAKTGGKFDFNCIQDLTYMDMFVSEVLRLWTPVVGMDRLCVKDYNLGRANKNATKDFILRKGEGLSIPTWSIHHNPEYYPEPYKFDPERFSEENKRNIKPFTYLPFGTGPRNCIGSRFALCEVKVMLYQLLQQIEVLPSDKTKVRAKLAKDTFNVKIEGGHWIRLKLRD
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
A5HKL6
A5HKL8
A4ZWB1
D0VY60
A5HKL7
L0N6I2
+ More
A5HKM0 H9J356 A4UU25 A5HMQ7 A3RIC2 A0A0K0YD31 A9Z0U9 A0A068ETV5 A0A2W1BPX7 A4KAE1 B6V7E2 A5HKL9 Q6RW46 L0N4K3 C6H0J5 H9J358 A3RKJ5 L0N4J7 L0N797 B6VEL8 D0FZ10 L0N725 X5DAZ5 A0A3G5EAT6 Q9U9C1 Q1KLD7 A0A0K8TUK1 A0A0L7L0K4 A0A0C5CGW0 Q5YJK0 A0A286MXM7 A0A0P0EW60 A0A3S2M2T6 Q9U9B9 A0A2A4JJX9 D2JLJ7 A0A0K8TUK8 A0A1V0D9E5 A0A385LJM0 A0A2A4JJV4 A0A2A4JDS7 A0A068F0Y7 A0A2A4JDS0 A0A1W5ZJN7 B2CPL1 A0A2A4JJD6 A0A2D1QTX7 A9Z0U8 A0A2W1BKQ5 V5JDC8 Q5XLI9 A0A248QJ18 Q4ZJH3 D2JLK7 A0A2W1BL52 B7TYK9 C9E6N0 A0A068EU83 B2CPL0 Q6UEI0 A0A194R3L1 A4KAE0 A0A194QDQ3 A0A385LJV6 A0A248QFE0 A0A194QJQ1 A0A2D1QTX0 A0A194QDY1 B5MF67 A0A068EXP4 Q7YZX4 A0A194R4E5 A0A3S2NVP0 B6ECI4 X5D4G9 A0A1V0D9E7 A0A385LJP1 A0A2D1QTX1 A0A212F932 A0A385LJT9 A0A0C5C1J2 A0A1W5ZJJ8 A0A346II56 A0A068ETV9 E5L8C3 A0A068EWN1 A0A2W1BR50 A0A248QEG5 A0A248QEG8 A0A0K0YD28 A0A068EUU6 W0FSV4 A0A2W1BSJ6
A5HKM0 H9J356 A4UU25 A5HMQ7 A3RIC2 A0A0K0YD31 A9Z0U9 A0A068ETV5 A0A2W1BPX7 A4KAE1 B6V7E2 A5HKL9 Q6RW46 L0N4K3 C6H0J5 H9J358 A3RKJ5 L0N4J7 L0N797 B6VEL8 D0FZ10 L0N725 X5DAZ5 A0A3G5EAT6 Q9U9C1 Q1KLD7 A0A0K8TUK1 A0A0L7L0K4 A0A0C5CGW0 Q5YJK0 A0A286MXM7 A0A0P0EW60 A0A3S2M2T6 Q9U9B9 A0A2A4JJX9 D2JLJ7 A0A0K8TUK8 A0A1V0D9E5 A0A385LJM0 A0A2A4JJV4 A0A2A4JDS7 A0A068F0Y7 A0A2A4JDS0 A0A1W5ZJN7 B2CPL1 A0A2A4JJD6 A0A2D1QTX7 A9Z0U8 A0A2W1BKQ5 V5JDC8 Q5XLI9 A0A248QJ18 Q4ZJH3 D2JLK7 A0A2W1BL52 B7TYK9 C9E6N0 A0A068EU83 B2CPL0 Q6UEI0 A0A194R3L1 A4KAE0 A0A194QDQ3 A0A385LJV6 A0A248QFE0 A0A194QJQ1 A0A2D1QTX0 A0A194QDY1 B5MF67 A0A068EXP4 Q7YZX4 A0A194R4E5 A0A3S2NVP0 B6ECI4 X5D4G9 A0A1V0D9E7 A0A385LJP1 A0A2D1QTX1 A0A212F932 A0A385LJT9 A0A0C5C1J2 A0A1W5ZJJ8 A0A346II56 A0A068ETV9 E5L8C3 A0A068EWN1 A0A2W1BR50 A0A248QEG5 A0A248QEG8 A0A0K0YD28 A0A068EUU6 W0FSV4 A0A2W1BSJ6
Pubmed
EMBL
EF535804
ABQ08706.1
EF535806
ABQ08708.2
BABH01036652
BABH01036653
+ More
EF491004 AK343209 ABP65279.1 BAM73893.1 AB500115 BAI49180.1 EF535805 ABQ08707.1 AK289300 BAM73827.1 EF535808 ABQ08710.1 BABH01036639 BABH01036640 BABH01036638 EF488994 ABP02071.1 EF542811 ABQ18318.1 EF415298 ABN71369.1 KR095603 AKS48891.1 EU327674 ABY47596.1 KM016750 AID54902.1 KZ149989 PZC75615.1 DQ788840 ABH09253.1 FJ265741 ACJ04711.1 EF535807 ABQ08709.1 AY487948 AAR37015.1 AK289302 BAM73829.1 FN421128 CAZ65619.1 BABH01036637 EF421989 ABO07439.1 AK289297 BAM73824.1 AK289298 BAM73825.1 FJ378716 ACJ05915.1 AB498807 BAI47532.1 AK289299 BAM73826.1 KF701145 AHW57315.1 MH001162 AYW35335.1 AF172279 AAD51036.1 DQ473438 ABF14737.1 GCVX01000068 JAI18162.1 JTDY01003828 KOB68945.1 KP001132 AJN91177.1 AY390260 AAR26518.1 MF684345 ASX93979.1 KR676343 ALJ30298.1 RSAL01000054 RVE50006.1 AF172281 AAD51038.1 NWSH01001322 PCG71702.1 GQ915313 ACZ97407.2 GCVX01000063 JAI18167.1 KY212055 ARA91619.1 MG793355 AYA28074.1 PCG71703.1 NWSH01001758 PCG70237.1 KM016753 AID54905.1 PCG70235.1 KY436738 ARI68318.1 EU541248 ACB30273.2 PCG71704.1 KY348420 ATP15901.1 EU327673 ABY47595.1 PZC75619.1 JX486677 AGI65181.1 AY753201 AAV28704.1 KX443436 ASO98011.1 DQ003275 AAY21809.1 GQ915323 ACZ97417.2 PZC75618.1 FJ416332 ACJ37388.1 GQ465040 ACV88722.1 KM016752 AID54904.1 EU541247 ACB30272.1 AY371318 AAQ73544.1 KQ460779 KPJ12393.1 DQ788839 ABH09252.1 KQ459144 KPJ03683.1 MG793354 AYA28073.1 KX443437 ASO98012.1 KPJ03681.1 KY348419 ATP15900.1 KPJ03682.1 AB381883 BAG71410.1 KJ671577 AID55429.1 AF525031 AAP80766.1 KPJ12394.1 RVE50007.1 FJ227150 ACI43222.1 KF701146 AHW57316.1 KY212054 ARA91618.1 MG793356 AYA28075.1 KY348418 ATP15899.1 AGBW02009651 OWR50238.1 MG793353 AYA28072.1 KP001131 AJN91176.1 KY436739 ARI68319.1 MH138270 AXP17211.1 KM016755 AID54907.1 HQ340240 KX443438 ADP55210.1 ASO98013.1 KJ671579 AID55431.1 PZC75617.1 KX443439 ASO98014.1 KX443440 ASO98015.1 KR095604 AKS48892.1 KJ671578 AID55430.1 KC832920 AHF45921.1 PZC75616.1
EF491004 AK343209 ABP65279.1 BAM73893.1 AB500115 BAI49180.1 EF535805 ABQ08707.1 AK289300 BAM73827.1 EF535808 ABQ08710.1 BABH01036639 BABH01036640 BABH01036638 EF488994 ABP02071.1 EF542811 ABQ18318.1 EF415298 ABN71369.1 KR095603 AKS48891.1 EU327674 ABY47596.1 KM016750 AID54902.1 KZ149989 PZC75615.1 DQ788840 ABH09253.1 FJ265741 ACJ04711.1 EF535807 ABQ08709.1 AY487948 AAR37015.1 AK289302 BAM73829.1 FN421128 CAZ65619.1 BABH01036637 EF421989 ABO07439.1 AK289297 BAM73824.1 AK289298 BAM73825.1 FJ378716 ACJ05915.1 AB498807 BAI47532.1 AK289299 BAM73826.1 KF701145 AHW57315.1 MH001162 AYW35335.1 AF172279 AAD51036.1 DQ473438 ABF14737.1 GCVX01000068 JAI18162.1 JTDY01003828 KOB68945.1 KP001132 AJN91177.1 AY390260 AAR26518.1 MF684345 ASX93979.1 KR676343 ALJ30298.1 RSAL01000054 RVE50006.1 AF172281 AAD51038.1 NWSH01001322 PCG71702.1 GQ915313 ACZ97407.2 GCVX01000063 JAI18167.1 KY212055 ARA91619.1 MG793355 AYA28074.1 PCG71703.1 NWSH01001758 PCG70237.1 KM016753 AID54905.1 PCG70235.1 KY436738 ARI68318.1 EU541248 ACB30273.2 PCG71704.1 KY348420 ATP15901.1 EU327673 ABY47595.1 PZC75619.1 JX486677 AGI65181.1 AY753201 AAV28704.1 KX443436 ASO98011.1 DQ003275 AAY21809.1 GQ915323 ACZ97417.2 PZC75618.1 FJ416332 ACJ37388.1 GQ465040 ACV88722.1 KM016752 AID54904.1 EU541247 ACB30272.1 AY371318 AAQ73544.1 KQ460779 KPJ12393.1 DQ788839 ABH09252.1 KQ459144 KPJ03683.1 MG793354 AYA28073.1 KX443437 ASO98012.1 KPJ03681.1 KY348419 ATP15900.1 KPJ03682.1 AB381883 BAG71410.1 KJ671577 AID55429.1 AF525031 AAP80766.1 KPJ12394.1 RVE50007.1 FJ227150 ACI43222.1 KF701146 AHW57316.1 KY212054 ARA91618.1 MG793356 AYA28075.1 KY348418 ATP15899.1 AGBW02009651 OWR50238.1 MG793353 AYA28072.1 KP001131 AJN91176.1 KY436739 ARI68319.1 MH138270 AXP17211.1 KM016755 AID54907.1 HQ340240 KX443438 ADP55210.1 ASO98013.1 KJ671579 AID55431.1 PZC75617.1 KX443439 ASO98014.1 KX443440 ASO98015.1 KR095604 AKS48892.1 KJ671578 AID55430.1 KC832920 AHF45921.1 PZC75616.1
Proteomes
PRIDE
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
A5HKL6
A5HKL8
A4ZWB1
D0VY60
A5HKL7
L0N6I2
+ More
A5HKM0 H9J356 A4UU25 A5HMQ7 A3RIC2 A0A0K0YD31 A9Z0U9 A0A068ETV5 A0A2W1BPX7 A4KAE1 B6V7E2 A5HKL9 Q6RW46 L0N4K3 C6H0J5 H9J358 A3RKJ5 L0N4J7 L0N797 B6VEL8 D0FZ10 L0N725 X5DAZ5 A0A3G5EAT6 Q9U9C1 Q1KLD7 A0A0K8TUK1 A0A0L7L0K4 A0A0C5CGW0 Q5YJK0 A0A286MXM7 A0A0P0EW60 A0A3S2M2T6 Q9U9B9 A0A2A4JJX9 D2JLJ7 A0A0K8TUK8 A0A1V0D9E5 A0A385LJM0 A0A2A4JJV4 A0A2A4JDS7 A0A068F0Y7 A0A2A4JDS0 A0A1W5ZJN7 B2CPL1 A0A2A4JJD6 A0A2D1QTX7 A9Z0U8 A0A2W1BKQ5 V5JDC8 Q5XLI9 A0A248QJ18 Q4ZJH3 D2JLK7 A0A2W1BL52 B7TYK9 C9E6N0 A0A068EU83 B2CPL0 Q6UEI0 A0A194R3L1 A4KAE0 A0A194QDQ3 A0A385LJV6 A0A248QFE0 A0A194QJQ1 A0A2D1QTX0 A0A194QDY1 B5MF67 A0A068EXP4 Q7YZX4 A0A194R4E5 A0A3S2NVP0 B6ECI4 X5D4G9 A0A1V0D9E7 A0A385LJP1 A0A2D1QTX1 A0A212F932 A0A385LJT9 A0A0C5C1J2 A0A1W5ZJJ8 A0A346II56 A0A068ETV9 E5L8C3 A0A068EWN1 A0A2W1BR50 A0A248QEG5 A0A248QEG8 A0A0K0YD28 A0A068EUU6 W0FSV4 A0A2W1BSJ6
A5HKM0 H9J356 A4UU25 A5HMQ7 A3RIC2 A0A0K0YD31 A9Z0U9 A0A068ETV5 A0A2W1BPX7 A4KAE1 B6V7E2 A5HKL9 Q6RW46 L0N4K3 C6H0J5 H9J358 A3RKJ5 L0N4J7 L0N797 B6VEL8 D0FZ10 L0N725 X5DAZ5 A0A3G5EAT6 Q9U9C1 Q1KLD7 A0A0K8TUK1 A0A0L7L0K4 A0A0C5CGW0 Q5YJK0 A0A286MXM7 A0A0P0EW60 A0A3S2M2T6 Q9U9B9 A0A2A4JJX9 D2JLJ7 A0A0K8TUK8 A0A1V0D9E5 A0A385LJM0 A0A2A4JJV4 A0A2A4JDS7 A0A068F0Y7 A0A2A4JDS0 A0A1W5ZJN7 B2CPL1 A0A2A4JJD6 A0A2D1QTX7 A9Z0U8 A0A2W1BKQ5 V5JDC8 Q5XLI9 A0A248QJ18 Q4ZJH3 D2JLK7 A0A2W1BL52 B7TYK9 C9E6N0 A0A068EU83 B2CPL0 Q6UEI0 A0A194R3L1 A4KAE0 A0A194QDQ3 A0A385LJV6 A0A248QFE0 A0A194QJQ1 A0A2D1QTX0 A0A194QDY1 B5MF67 A0A068EXP4 Q7YZX4 A0A194R4E5 A0A3S2NVP0 B6ECI4 X5D4G9 A0A1V0D9E7 A0A385LJP1 A0A2D1QTX1 A0A212F932 A0A385LJT9 A0A0C5C1J2 A0A1W5ZJJ8 A0A346II56 A0A068ETV9 E5L8C3 A0A068EWN1 A0A2W1BR50 A0A248QEG5 A0A248QEG8 A0A0K0YD28 A0A068EUU6 W0FSV4 A0A2W1BSJ6
PDB
6MA7
E-value=3.12318e-59,
Score=580
Ontologies
KEGG
GO
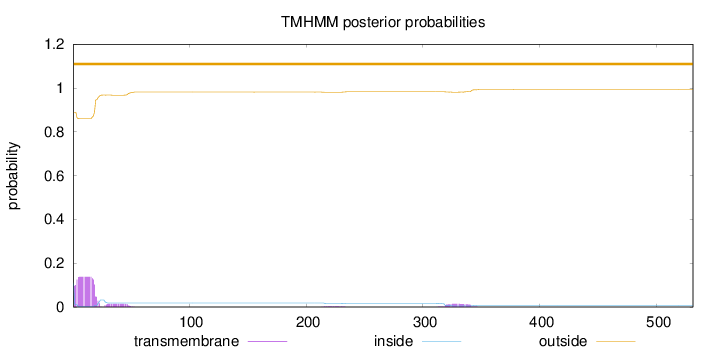
Topology
SignalP
Position: 1 - 23,
Likelihood: 0.669546

Length:
531
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.12969000000001
Exp number, first 60 AAs:
2.76087
Total prob of N-in:
0.11034
outside
1 - 531
Population Genetic Test Statistics
Pi
205.859876
Theta
171.731515
Tajima's D
1.06407
CLR
0.147759
CSRT
0.67921603919804
Interpretation
Uncertain
