Gene
KWMTBOMO10621
Pre Gene Modal
BGIBMGA003944
Annotation
Cytochrome_P450_9e2_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.412
Sequence
CDS
ATGATTATCATTATATGGACATTAGCTATAGGTTTGGCTTTCTTACTGTACCTGAAGCAGATATATTGCTACTTCAGCAAGCATGAAATCAAGAGCATAACCCCGCTTCCGATACTGGGCAACATGGGCAAGATTGTCTTCAAGATTAACCATTTTGTTGATGATATTTCTCAATTGTACAACAAATTTCCAGAAGAAAGATTCGTTGGTAGATATGAGTTTGTAAATCCGGTGATCTACATCAGAGACATTGAGATCGTAAAGAGAATCACAATTAAAGACTTTGAACATTTCCTGGATCATCGCACGATCGTCAATGAGGAAACCGATCCCATTTTCGGAAGGAATCTGTTTTCTTTGAAAGGCCAGGAATGGAAAGATATGCGATCCACTTTATCGCCAGCCTTCACCAGCTCCAAGATGAAGCTTATGATGCCCTTAATAGTCGAAGTTGGAGAGCAGATGATCAACGCGTTAAAAAAGAACATTAAAAACTCTGGTGTCGGATACGTGGACATAGACACTAAAGACCTGACCACTAGATACGCCAACGATGTGATCGCCTCCTGCGCCTTCGGTCTGAAGGTCGACTCCCTCACCGAAGAGAACAACCAATTCTACGCGATGGGGAAAGCCGCTTCGAATTTCAGTTTCAAACAGATCCTACTCTTGTTGGGATTTATCTCATTTCCTAAGATGATGAAGATGACGAAATTCACGCTGTTTTCGGAAGAAACGTCCGGATTCTTTAAAGAGCTGATAATGGGTACCATGAAAGACAGAGAAATGCGGAAAATCATCAGACCAGACATGATCCATCTACTCATGGAAGCTAAGAAAGGAAAATTAGTCCACGATGATAAATCCAGCAAAGACACAGATGCCGGATTCGCCACGGTAGAAGAATCTGCTGTTGGCAAGAAACAAATCGATAGAGTCTGGACTGATGACGACATCATCGCTCAAGCAGTTCTTTTCTTCGTTGCTGGTTTTGAAACCGTTTCATCGGCAATGACGTTCCTCCTTCACGAATTGGCTCTCAACCCTGAAGTACAAGACAAATTGGTTGAAGAAATCAAGGAGAACAAGGAGAGGAACAATGGAAAATTTGACTACAACTCCATACAGAATATGGTGTATCTGGATATGGTTGTTTCAGAACTCCTAAGGCTGTGGCCGCCAGGTGTCTCCATGGATAGAATCTGCGTTCAAGACTACAACTTGGGGAAACCAAATGACAAGGCCAAAAGAGACTTCATACTTCGCAAAGGTACGGGTGTAGCTATTCCCGTATGGGCTTTCCACCGAAACCCGGAATTCTTCCCGGACCCCCTGAAATTCGATCCCGAAAGGTTCTCAGAGGAGAACAAACACAACATCAAGCCTTTCGCGTATCTACCATTTGGAGTGGGACCTAGGAATTGTATAGGTTCTAGGTTCGCTTTATGCGAAGTCAAAGTGATGGCATACCAGCTACTCCAGCATATGGAGATATCTCCGTGCGAGAAGACGTGTATCCCCTCGAAGCTCAGCAAGGAAACCTTCAACCTTCGTCTTGAAGGTGGTAAAATGATCATCATCATATGGACATTAGTGATAGGTTTGGCTCTCTTACTGTACCTGAAGCAGACATATTCCTACTTCAGCAAGCATGAAATCAAGAGCGTAACTCCGCTTCCGATACTGGGCAACATGGGCAAGATCGTCTTTAAATTTAACCATTTAGTAGATGACATTTCGCAATTGTACAATAATTTTCCAGAAGAAAGATTCGTTGGTAGATACGAGTTTGTAAATCCGGTGATCTACATCAGAGACATTGAGATCGTAAAGAGAATCACAATTAAAGACTTTGAACATTTCCTGGATCATCGCACGATCGTCAATGAGGATTCCGATCCGATGTTTGGAAGGAATCTGTTTTCATTGAAAGGCCAGGAATGGAAAGATATGCGATCCACTTTATCACCAGCCTTCACTAGTTCCAAGATGAAGCTTATGATGCCCTTAATAGTCGAAGTGGGAGAGCAGATGATCAACGCCATAAAAGAGAATATTAAAAACTCTGGTGTCGGATACGTGGACATAGACACTAAAGACCTGACCACTAGATACGCCAACGATGTGATCGCGTCCTGCGCCTTCGGTCTGAAGGTCGACTCACTCACCGAGGAGAACAACAGATTCTACGCGATGGGGAAAGCCGCTACGAATTTCAGTTTCAAACAGATCCTAATGTTGTTGGGATTTATTTCATTTCCTAAGTTGATGAAGATGACGAAATTCAGACTGTTTTCGGAAGAAACGTCCGGATTCTTTAAGGAGCTGATAATGGGCACCATGAAGGACAGGGAAATGCGGAAAATCATCAGACCAGACATGATACACTTACTCATGGAAGCTAAGAAAGGAAAATTAGTTCACGACAATAAATCAAGCAAAGACACAGATGCCGGATTCGCCACGGTAGAAGAATCTGCTGTTGGCAAAAAACAAATCGATAGAGTCTGGACTGATGACGACATCATCGCTCAAGCAGTTCTTTTCTTCATTGCTGGTTTTGAGACCGTTTCATCGGCAATGACGTTCCTCCTTCACGAATTAGCTCTCAACCCTGAGGTACAAGAAAAGCTGGTTGAAGAAATCAAGGAGAACAAGGAGAGGAACAATGGGAAATTTGACTATAACTCCATACAGAATATGGCGTATTTGGATATGGTTGTTTCAGAACTCCTAAGGCTGTGGCCGCCAGCTGTCTCCATGGATAGAATCTGCGTTCAAGACTACAACTTGGGGAAACCAAATGACAAGGCCAAAAGAGACTTCATACTTCGCAAAGGTACGGGTGTAGCTATTCCCGTATGGGCTTTTCACCGAAACCCGGAATTCTTCCCGGACCCCCAGAAATTCGATCCCGAAAGGTTCTCAGAGGAGAACAAACACAACATCAAGCCTTTCACGTATCTACCCTTTGGTGTGGGACCTAGGAACTGTATAGGTTCCAGGTTTGCTTTATGCGAAGTCAAAGTGATGGCATACCAGCTACTCCAGCATATGGAGATATCTCCGTGCGAGAAGACGTGCATCCCCTCGAAGCTCAGCAAGGAGATCTTCAACCTTCGTCTTGAAGGTGGTCATTGGGTCAGGCTAAAGATTAGAGATTGA
Protein
MIIIIWTLAIGLAFLLYLKQIYCYFSKHEIKSITPLPILGNMGKIVFKINHFVDDISQLYNKFPEERFVGRYEFVNPVIYIRDIEIVKRITIKDFEHFLDHRTIVNEETDPIFGRNLFSLKGQEWKDMRSTLSPAFTSSKMKLMMPLIVEVGEQMINALKKNIKNSGVGYVDIDTKDLTTRYANDVIASCAFGLKVDSLTEENNQFYAMGKAASNFSFKQILLLLGFISFPKMMKMTKFTLFSEETSGFFKELIMGTMKDREMRKIIRPDMIHLLMEAKKGKLVHDDKSSKDTDAGFATVEESAVGKKQIDRVWTDDDIIAQAVLFFVAGFETVSSAMTFLLHELALNPEVQDKLVEEIKENKERNNGKFDYNSIQNMVYLDMVVSELLRLWPPGVSMDRICVQDYNLGKPNDKAKRDFILRKGTGVAIPVWAFHRNPEFFPDPLKFDPERFSEENKHNIKPFAYLPFGVGPRNCIGSRFALCEVKVMAYQLLQHMEISPCEKTCIPSKLSKETFNLRLEGGKMIIIIWTLVIGLALLLYLKQTYSYFSKHEIKSVTPLPILGNMGKIVFKFNHLVDDISQLYNNFPEERFVGRYEFVNPVIYIRDIEIVKRITIKDFEHFLDHRTIVNEDSDPMFGRNLFSLKGQEWKDMRSTLSPAFTSSKMKLMMPLIVEVGEQMINAIKENIKNSGVGYVDIDTKDLTTRYANDVIASCAFGLKVDSLTEENNRFYAMGKAATNFSFKQILMLLGFISFPKLMKMTKFRLFSEETSGFFKELIMGTMKDREMRKIIRPDMIHLLMEAKKGKLVHDNKSSKDTDAGFATVEESAVGKKQIDRVWTDDDIIAQAVLFFIAGFETVSSAMTFLLHELALNPEVQEKLVEEIKENKERNNGKFDYNSIQNMAYLDMVVSELLRLWPPAVSMDRICVQDYNLGKPNDKAKRDFILRKGTGVAIPVWAFHRNPEFFPDPQKFDPERFSEENKHNIKPFTYLPFGVGPRNCIGSRFALCEVKVMAYQLLQHMEISPCEKTCIPSKLSKEIFNLRLEGGHWVRLKIRD
Summary
Similarity
Belongs to the cytochrome P450 family.
Uniprot
Pubmed
EMBL
KQ459144
KPJ03682.1
AGBW02009651
OWR50238.1
AJWK01032956
JXUM01009174
+ More
KQ560277 KXJ83348.1 KB632166 ERL89339.1 KK852611 KDR20273.1 CVRI01000064 CRL05076.1 KQ981253 KYN44186.1 KK107147 EZA57483.1 KQ981042 KYN10063.1 KK107154 EZA56952.1 KQ979548 KYN21066.1 QOIP01000013 RLU15532.1 KQ977444 KYN02645.1 KQ971363 KYB25692.1 GL888387 EGI61800.1 GL762454 EFZ21470.1 KYB25693.1
KQ560277 KXJ83348.1 KB632166 ERL89339.1 KK852611 KDR20273.1 CVRI01000064 CRL05076.1 KQ981253 KYN44186.1 KK107147 EZA57483.1 KQ981042 KYN10063.1 KK107154 EZA56952.1 KQ979548 KYN21066.1 QOIP01000013 RLU15532.1 KQ977444 KYN02645.1 KQ971363 KYB25692.1 GL888387 EGI61800.1 GL762454 EFZ21470.1 KYB25693.1
Proteomes
Pfam
PF00067 p450
Interpro
SUPFAM
SSF48264
SSF48264
Gene 3D
ProteinModelPortal
PDB
6MA7
E-value=2.77703e-67,
Score=652
Ontologies
GO
Topology
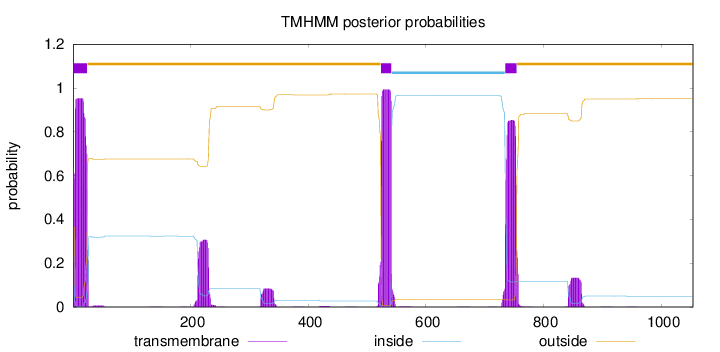
Length:
1054
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
67.25804
Exp number, first 60 AAs:
20.85668
Total prob of N-in:
0.63632
POSSIBLE N-term signal
sequence
inside
1 - 1
TMhelix
2 - 24
outside
25 - 523
TMhelix
524 - 541
inside
542 - 734
TMhelix
735 - 754
outside
755 - 1054
Population Genetic Test Statistics
Pi
209.27108
Theta
173.970703
Tajima's D
0.936119
CLR
0.280102
CSRT
0.644467776611169
Interpretation
Uncertain
