Gene
KWMTBOMO10611
Pre Gene Modal
BGIBMGA003951
Annotation
hypothetical_protein_KGM_05135_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.074 Nuclear Reliability : 1.191 PlasmaMembrane Reliability : 1.005
Sequence
CDS
ATGCACGCGGTGGGACTACACTCCGCGCTCGCCATCCGCCGCGAGCGGAAGCGCCGCGCCGAGCAGCAGCGCGCTCGCGAGCGCCGCTACTCGCTGCAGTCCTCCGAGTCCGGCCTCACCAGCCCGCACTGCTCCACCGGCAGCTTGGAGAGGCGCCGCTACCGGAACACCAGGGCCGCCGAAAATGAAGTCGTCTCCTCCGTTGGCATGCTGCACCTCGGTGTCGTCTTCCTCGTACTCGGCGTCTTCCTGGTCGCCAGCGGATGGTTGCCTGACGACGTCACCTCTTGGAGTAGTATCGGCTCCGTCAGCTGGTTCAACGAGCTGGTCTGCTCGGGTTTGTTCGCGCTTGGGATCGGGATATTTCTGATCGCTCTGCACAAGTACCTCACCAAGAGCGAGGAGGAGGCGCTAGAGGACTATGTGCAGAGACAGCTAACGAGGTCACGATCAGGGCACCGGCTGGAGAGGGACGCGGAGACAGGCGGCATGCACACGAAGAACGCGCGGCGAGCCCGCGCCACCGAGGTGCTGCCGGATGCGACCTCCGAGACGTACACGCAGCCCCCCGAGCGCGCGCACGGGTTCGTCAATGCTCTGGCGGTGAACGGCGACGCTCTGCGGGAGATGCCGCTCGAGCAAATCGCGGAGGAGGAGGTATCGGTGACGTCTGAGTGCGACCGGCGGCTCGGCAAGTTCAACAAGGACACGTACTCGACCCCGTCCGTGGCGCCCAGCCTGAGCCCGGGCTCGCCGTCCGACACGCGGGAGCTGCTCTCCGACGGGCGCTATATGGTCATGTCGCGCATATGA
Protein
MHAVGLHSALAIRRERKRRAEQQRARERRYSLQSSESGLTSPHCSTGSLERRRYRNTRAAENEVVSSVGMLHLGVVFLVLGVFLVASGWLPDDVTSWSSIGSVSWFNELVCSGLFALGIGIFLIALHKYLTKSEEEALEDYVQRQLTRSRSGHRLERDAETGGMHTKNARRARATEVLPDATSETYTQPPERAHGFVNALAVNGDALREMPLEQIAEEEVSVTSECDRRLGKFNKDTYSTPSVAPSLSPGSPSDTRELLSDGRYMVMSRI
Summary
Uniprot
H9J364
A0A212FM24
A0A2H1VNY0
A0A2W1BRL0
A0A0N1IN98
A0A194R594
+ More
A0A0J7L5R7 A0A195F6Q5 A0A158NJG5 E2AWY2 A0A151IDZ0 A0A088A8N3 A0A151HXM2 A0A2P8YZS7 A0A151WI72 A0A0C9RE04 A0A0M9A7H1 K7J9L3 A0A2J7QAN3 A0A336M4Z4 F4WDQ7 E2BQL2 E9I951 A0A084WU31 A0A182J5E7 A0A182NBR1 A0A182Y0N4 Q16TA0 A0A182QDS7 A0A336LHC7 A0A182KU35 A0A182LX21 A0A182USQ7 A0A182WGI7 A0NFU4 A0A182R3T3 W5JGX4 A0A067R9C7 A0A182K8B9 A0A182TFF6 A0A026W3I8 A0A0L7R0V4 A0A1B0CE29 A0A0K8SLK8 A0A1A9WXD9 A0A2R7WHK7 A0A1A9YJV8 A0A1A9UYR3 A0A1A9Z441 A0A1B6M9I7 D7EKH3 A0A0P8XKP0 A0A1W4X2C2 N6SZB2 A0A1W4VUT0 U4TUK1 A0A0Q9X6C4 A0A1I8NWR3 A0A0Q5WQJ1 E0VHH4 A8Y4V5 A0A0R1E780 A0A1W4W618 A0A0A1WUC0 A0A182F0X4 A0A0A1WUH8 A0A0K8UK91 A0A232EU29 A0A034WG93 W8B9Q6 A0A0K8VTG9 A0A0R3NVY7 A0A0L0CMC2 A0A3B0KIJ8
A0A0J7L5R7 A0A195F6Q5 A0A158NJG5 E2AWY2 A0A151IDZ0 A0A088A8N3 A0A151HXM2 A0A2P8YZS7 A0A151WI72 A0A0C9RE04 A0A0M9A7H1 K7J9L3 A0A2J7QAN3 A0A336M4Z4 F4WDQ7 E2BQL2 E9I951 A0A084WU31 A0A182J5E7 A0A182NBR1 A0A182Y0N4 Q16TA0 A0A182QDS7 A0A336LHC7 A0A182KU35 A0A182LX21 A0A182USQ7 A0A182WGI7 A0NFU4 A0A182R3T3 W5JGX4 A0A067R9C7 A0A182K8B9 A0A182TFF6 A0A026W3I8 A0A0L7R0V4 A0A1B0CE29 A0A0K8SLK8 A0A1A9WXD9 A0A2R7WHK7 A0A1A9YJV8 A0A1A9UYR3 A0A1A9Z441 A0A1B6M9I7 D7EKH3 A0A0P8XKP0 A0A1W4X2C2 N6SZB2 A0A1W4VUT0 U4TUK1 A0A0Q9X6C4 A0A1I8NWR3 A0A0Q5WQJ1 E0VHH4 A8Y4V5 A0A0R1E780 A0A1W4W618 A0A0A1WUC0 A0A182F0X4 A0A0A1WUH8 A0A0K8UK91 A0A232EU29 A0A034WG93 W8B9Q6 A0A0K8VTG9 A0A0R3NVY7 A0A0L0CMC2 A0A3B0KIJ8
Pubmed
19121390
22118469
28756777
26354079
21347285
20798317
+ More
29403074 20075255 21719571 21282665 24438588 25244985 17510324 20966253 12364791 14747013 17210077 20920257 23761445 24845553 24508170 18362917 19820115 17994087 23537049 18057021 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 28648823 25348373 24495485 15632085 26108605
29403074 20075255 21719571 21282665 24438588 25244985 17510324 20966253 12364791 14747013 17210077 20920257 23761445 24845553 24508170 18362917 19820115 17994087 23537049 18057021 20566863 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 28648823 25348373 24495485 15632085 26108605
EMBL
BABH01036614
AGBW02007658
OWR54792.1
ODYU01003575
SOQ42508.1
KZ150001
+ More
PZC75306.1 KQ458903 KPJ04567.1 KQ460779 KPJ12410.1 LBMM01000647 KMQ97898.1 KQ981798 KYN35739.1 ADTU01017868 GL443425 EFN62102.1 KQ977901 KYM98918.1 KQ976748 KYM75309.1 PYGN01000263 PSN49751.1 KQ983097 KYQ47490.1 GBYB01014779 JAG84546.1 KQ435732 KOX77503.1 AAZX01002604 NEVH01016330 PNF25619.1 UFQT01000171 SSX21088.1 GL888090 EGI67670.1 GL449769 EFN82070.1 GL761703 EFZ22902.1 ATLV01026995 KE525421 KFB53725.1 AXCP01000261 AXCP01000262 CH477657 EAT37689.1 AXCN02000684 UFQS01002095 UFQT01002095 SSX13222.1 SSX32661.1 AXCM01001089 AAAB01008980 EAU76033.2 ADMH02001427 ETN62573.1 KK852804 KDR16204.1 KK107503 EZA49594.1 KQ414669 KOC64478.1 AJWK01008529 AJWK01008530 AJWK01008531 GBRD01011686 JAG54138.1 KK854837 PTY19134.1 GEBQ01007398 JAT32579.1 KQ971357 EFA13155.2 CH902628 KPU75347.1 APGK01058729 KB741291 ENN70568.1 KB631579 ERL84442.1 CH933807 KRG03813.1 KRG03814.1 KRG03815.1 KRG03816.1 CH954177 KQS71064.1 DS235170 EEB12830.1 BT150085 AE013599 AGK45258.1 EAL24627.2 CH891641 KRK05093.1 KRK05094.1 GBXI01012269 GBXI01005373 JAD02023.1 JAD08919.1 GBXI01011785 JAD02507.1 GDHF01025182 JAI27132.1 NNAY01002181 OXU21858.1 GAKP01004346 JAC54606.1 GAMC01008611 GAMC01008610 JAB97945.1 GDHF01010118 JAI42196.1 CH475486 KRT05156.1 JRES01000300 KNC32619.1 OUUW01000010 SPP85586.1
PZC75306.1 KQ458903 KPJ04567.1 KQ460779 KPJ12410.1 LBMM01000647 KMQ97898.1 KQ981798 KYN35739.1 ADTU01017868 GL443425 EFN62102.1 KQ977901 KYM98918.1 KQ976748 KYM75309.1 PYGN01000263 PSN49751.1 KQ983097 KYQ47490.1 GBYB01014779 JAG84546.1 KQ435732 KOX77503.1 AAZX01002604 NEVH01016330 PNF25619.1 UFQT01000171 SSX21088.1 GL888090 EGI67670.1 GL449769 EFN82070.1 GL761703 EFZ22902.1 ATLV01026995 KE525421 KFB53725.1 AXCP01000261 AXCP01000262 CH477657 EAT37689.1 AXCN02000684 UFQS01002095 UFQT01002095 SSX13222.1 SSX32661.1 AXCM01001089 AAAB01008980 EAU76033.2 ADMH02001427 ETN62573.1 KK852804 KDR16204.1 KK107503 EZA49594.1 KQ414669 KOC64478.1 AJWK01008529 AJWK01008530 AJWK01008531 GBRD01011686 JAG54138.1 KK854837 PTY19134.1 GEBQ01007398 JAT32579.1 KQ971357 EFA13155.2 CH902628 KPU75347.1 APGK01058729 KB741291 ENN70568.1 KB631579 ERL84442.1 CH933807 KRG03813.1 KRG03814.1 KRG03815.1 KRG03816.1 CH954177 KQS71064.1 DS235170 EEB12830.1 BT150085 AE013599 AGK45258.1 EAL24627.2 CH891641 KRK05093.1 KRK05094.1 GBXI01012269 GBXI01005373 JAD02023.1 JAD08919.1 GBXI01011785 JAD02507.1 GDHF01025182 JAI27132.1 NNAY01002181 OXU21858.1 GAKP01004346 JAC54606.1 GAMC01008611 GAMC01008610 JAB97945.1 GDHF01010118 JAI42196.1 CH475486 KRT05156.1 JRES01000300 KNC32619.1 OUUW01000010 SPP85586.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000036403
UP000078541
+ More
UP000005205 UP000000311 UP000078542 UP000005203 UP000078540 UP000245037 UP000075809 UP000053105 UP000002358 UP000235965 UP000007755 UP000008237 UP000030765 UP000075880 UP000075884 UP000076408 UP000008820 UP000075886 UP000075882 UP000075883 UP000075903 UP000075920 UP000007062 UP000075900 UP000000673 UP000027135 UP000075881 UP000075902 UP000053097 UP000053825 UP000092461 UP000091820 UP000092443 UP000078200 UP000092445 UP000007266 UP000007801 UP000192223 UP000019118 UP000192221 UP000030742 UP000009192 UP000095300 UP000008711 UP000009046 UP000000803 UP000002282 UP000069272 UP000215335 UP000001819 UP000037069 UP000268350
UP000005205 UP000000311 UP000078542 UP000005203 UP000078540 UP000245037 UP000075809 UP000053105 UP000002358 UP000235965 UP000007755 UP000008237 UP000030765 UP000075880 UP000075884 UP000076408 UP000008820 UP000075886 UP000075882 UP000075883 UP000075903 UP000075920 UP000007062 UP000075900 UP000000673 UP000027135 UP000075881 UP000075902 UP000053097 UP000053825 UP000092461 UP000091820 UP000092443 UP000078200 UP000092445 UP000007266 UP000007801 UP000192223 UP000019118 UP000192221 UP000030742 UP000009192 UP000095300 UP000008711 UP000009046 UP000000803 UP000002282 UP000069272 UP000215335 UP000001819 UP000037069 UP000268350
Pfam
PF00075 RNase_H
Interpro
Gene 3D
ProteinModelPortal
H9J364
A0A212FM24
A0A2H1VNY0
A0A2W1BRL0
A0A0N1IN98
A0A194R594
+ More
A0A0J7L5R7 A0A195F6Q5 A0A158NJG5 E2AWY2 A0A151IDZ0 A0A088A8N3 A0A151HXM2 A0A2P8YZS7 A0A151WI72 A0A0C9RE04 A0A0M9A7H1 K7J9L3 A0A2J7QAN3 A0A336M4Z4 F4WDQ7 E2BQL2 E9I951 A0A084WU31 A0A182J5E7 A0A182NBR1 A0A182Y0N4 Q16TA0 A0A182QDS7 A0A336LHC7 A0A182KU35 A0A182LX21 A0A182USQ7 A0A182WGI7 A0NFU4 A0A182R3T3 W5JGX4 A0A067R9C7 A0A182K8B9 A0A182TFF6 A0A026W3I8 A0A0L7R0V4 A0A1B0CE29 A0A0K8SLK8 A0A1A9WXD9 A0A2R7WHK7 A0A1A9YJV8 A0A1A9UYR3 A0A1A9Z441 A0A1B6M9I7 D7EKH3 A0A0P8XKP0 A0A1W4X2C2 N6SZB2 A0A1W4VUT0 U4TUK1 A0A0Q9X6C4 A0A1I8NWR3 A0A0Q5WQJ1 E0VHH4 A8Y4V5 A0A0R1E780 A0A1W4W618 A0A0A1WUC0 A0A182F0X4 A0A0A1WUH8 A0A0K8UK91 A0A232EU29 A0A034WG93 W8B9Q6 A0A0K8VTG9 A0A0R3NVY7 A0A0L0CMC2 A0A3B0KIJ8
A0A0J7L5R7 A0A195F6Q5 A0A158NJG5 E2AWY2 A0A151IDZ0 A0A088A8N3 A0A151HXM2 A0A2P8YZS7 A0A151WI72 A0A0C9RE04 A0A0M9A7H1 K7J9L3 A0A2J7QAN3 A0A336M4Z4 F4WDQ7 E2BQL2 E9I951 A0A084WU31 A0A182J5E7 A0A182NBR1 A0A182Y0N4 Q16TA0 A0A182QDS7 A0A336LHC7 A0A182KU35 A0A182LX21 A0A182USQ7 A0A182WGI7 A0NFU4 A0A182R3T3 W5JGX4 A0A067R9C7 A0A182K8B9 A0A182TFF6 A0A026W3I8 A0A0L7R0V4 A0A1B0CE29 A0A0K8SLK8 A0A1A9WXD9 A0A2R7WHK7 A0A1A9YJV8 A0A1A9UYR3 A0A1A9Z441 A0A1B6M9I7 D7EKH3 A0A0P8XKP0 A0A1W4X2C2 N6SZB2 A0A1W4VUT0 U4TUK1 A0A0Q9X6C4 A0A1I8NWR3 A0A0Q5WQJ1 E0VHH4 A8Y4V5 A0A0R1E780 A0A1W4W618 A0A0A1WUC0 A0A182F0X4 A0A0A1WUH8 A0A0K8UK91 A0A232EU29 A0A034WG93 W8B9Q6 A0A0K8VTG9 A0A0R3NVY7 A0A0L0CMC2 A0A3B0KIJ8
Ontologies
GO
PANTHER
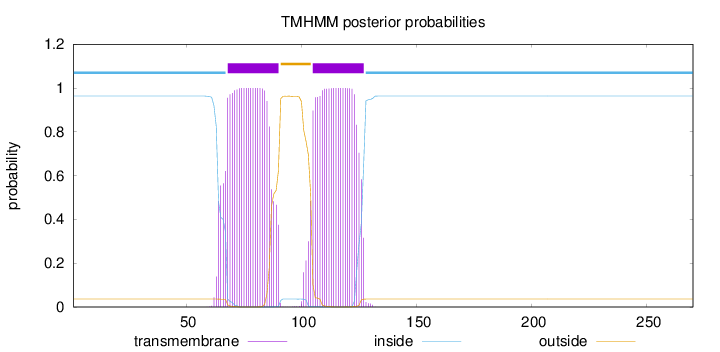
Topology
Length:
270
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
45.3353
Exp number, first 60 AAs:
0.00328
Total prob of N-in:
0.96329
inside
1 - 67
TMhelix
68 - 90
outside
91 - 104
TMhelix
105 - 127
inside
128 - 270
Population Genetic Test Statistics
Pi
169.752676
Theta
127.734042
Tajima's D
1.156778
CLR
73.406656
CSRT
0.701414929253537
Interpretation
Uncertain
